import pandas as pd
import pickle
from myst_nb import glue
# Define variables with glue here
scores_df = pd.read_csv("../results/tables/model_comparison_results.csv",index_col=0)
glue("dt_test_score", scores_df['test_score'].values[1], display = False)
glue("lr_test_score", scores_df['test_score'].values[2], display = False)
glue("optimized_knn_test_score", scores_df['test_score'].values[3],display=False)
glue("dummy_score", scores_df['test_score'].values[0], display = False)
glue("model_score_table", scores_df, display=False)
coef_df = pd.read_csv("../results/tables/feature_importances.csv",index_col=0).sort_values(by='coefficients', key = abs, ascending = False).round(2).query("features != 'ID'")
glue("coef_table", coef_df, display=False)
glue("highest_importance_coeff", coef_df['coefficients'].values[0],display=False)
confusion_matrix_dt = pd.read_csv("../results/tables/confusion_matrix_DT.csv",index_col=0)
glue("false_negative", confusion_matrix_dt.iloc[1,0],display=False)
glue("false_positive", confusion_matrix_dt.iloc[0,1],display=False)
best_params = pd.read_csv("../results/tables/best_hyperparams.csv",index_col=0)
glue("knn_bestparam", best_params.loc[:, "n_neighbours"][0],display=False)
glue("tree_bestparam", best_params.loc[:, "max_depth"][0],display=False)
Diabetes Classification Model#
Authors: Angela Chen, Ella Hein, Scout McKee, Sharon Voon
Summary#
This project endeavors to develop a predictive classification model for ascertaining an individual’s diabetic status, while comparing the efficiency of optimized decision tree and optimized k-nearest neighbours (k-nn) algorithms. The dataset used in this analysis is collected through the Behavioral Risk Factor Surveillance System (BFRSS) by the Centers for Disease Control and Prevention (CDC) for the year 2015. Notably, the primary determinant influencing the prediction is identified as the feature General Health Factor (1 being excellent and 5 being poor), displaying a coefficient of 0.55 as revealed by the logistic regression model. The optimized decision tree model demonstrates a test score of 0.852 (+/- 0.001),while the optimized k-nn model yields a test score of 0.847 (+/- 0.000). Both of the test scores are relatively close to the validation score which shows that the model will generalized well to unseen data, however, there is still room for improvement in the test score.
Our final classsifer is chosen to be the Desicion Tree model as it has both the highest test and train score in comparison with the optimized k-nn model. However, from the confusion matrix, we can see that our model has made 11179 type II error, which predicted the individual as non-diabetic when they are actually diabetic. On the other hand, it has also made 784 type I error, which predicted the individual as diabetic when they are actually non-diabetic. In our case, both type II error and type I error is equivalently important as injecting insulin in a non-diabetic individual can be fatal and vice versa. Hence, we chose f1 score as our scoring matrix which is a hormonic balance between both type I and type II errors.
From this result, we can also see that there is a huge class imbalance in between diabetic and non-diabetic class, which will be taken into account in the preprocessor in the next version of this analysis.
Continued efforts in these areas will contribute to the model’s robustness and reliability, making it more suitable for deployment in a clinical setting.
Introduction#
Diabetes mellitus, commonly referred to as diabetes is a disease which impacts the body’s control of blood glucose levels [Sapra and Bhandari, 2023]. It is important to note that there are different types of diabetes, although we do not explore this discrepancy in this project [Sapra and Bhandari, 2023]. Diabetes is a manageable disease thanks to the discovery of insulin in 1922. Globally, 1 in 11 adults have diabetes [Sapra and Bhandari, 2023]. As such, understanding the factors which are strongly related to diabetes can be important for researchers studying how to better prevent or manage the disease. In this project, we create several machine learning models to predict diabetes in a patient and evaluate the success of these models. We also explore the coefficients of a logistic regression model to better understand the factors which are associated with diabetes.
Methods#
Data#
The dataset used in this project is a collection of the Centers for Disease Control and Prevention (CDC) diabetes health indicators collected as a response to the CDC’s BRFSS2015 survey. The data were sourced from the UCI Machine Learning Repository [Burrows et al., 2017] which can be found here. The file specifically used from this repository for this analysis includes 70, 692 survey responses from which 50% of the respondents recorded having either prediabetes or diabetes. Each row in the dataset represents a recorded survey response including whether or not the responded has diabetes or prediabetes, and a collection of 21 other diabetes health indicators identified by the CDC.
Analysis#
In our efforts to determine the best model for classifying a patient with diabetes or prediabetes as opposed to no diabetes or prediabetes, we performed hyperparamter optimization on both a knn model and a decision tree model. We also explored a logistic regression model to gain insight into which features may contribute most to a classification of diabetes. All features from the original dataset were included in each model. In all cases, the data were split into training and testing datasets, with 65% of the data designated as training and 35% as testing. The data was preprocessesed such that all continuous (non-binary) variables were scaled using a scikit-learn’s StandardScaler function. Model performance was tested using a 10 - fold cross validation score. Feature importance was investigated using the coefficients generated by the logistic regression algorithm. The k-nn algorithm’s hyperparameter K was optimized using the F1 score as the classification metric. Python programming (Van Rossum and Drake 2009) was used for all analysis. The following Python packages were used for this analysis: Pandas [Wes McKinney, 2010], altair [VanderPlas et al., 2018], and scikit-learn [Pedregosa et al., 2011].
Results & Discussion#
Before we begin building our models for comparison, we plotted the histogram distribution of each feature in the dataset to have an insight on the features, using 0 and 1 representing the non-diabetic class(blue) and the diabetic class(orange) respectively. It is seen that for all the features, there is a significant overlapping in between the classes as most of the features are binary features. However, their max and mean values for each feature seems to be different, thus, we decided to include all the features in our model training.
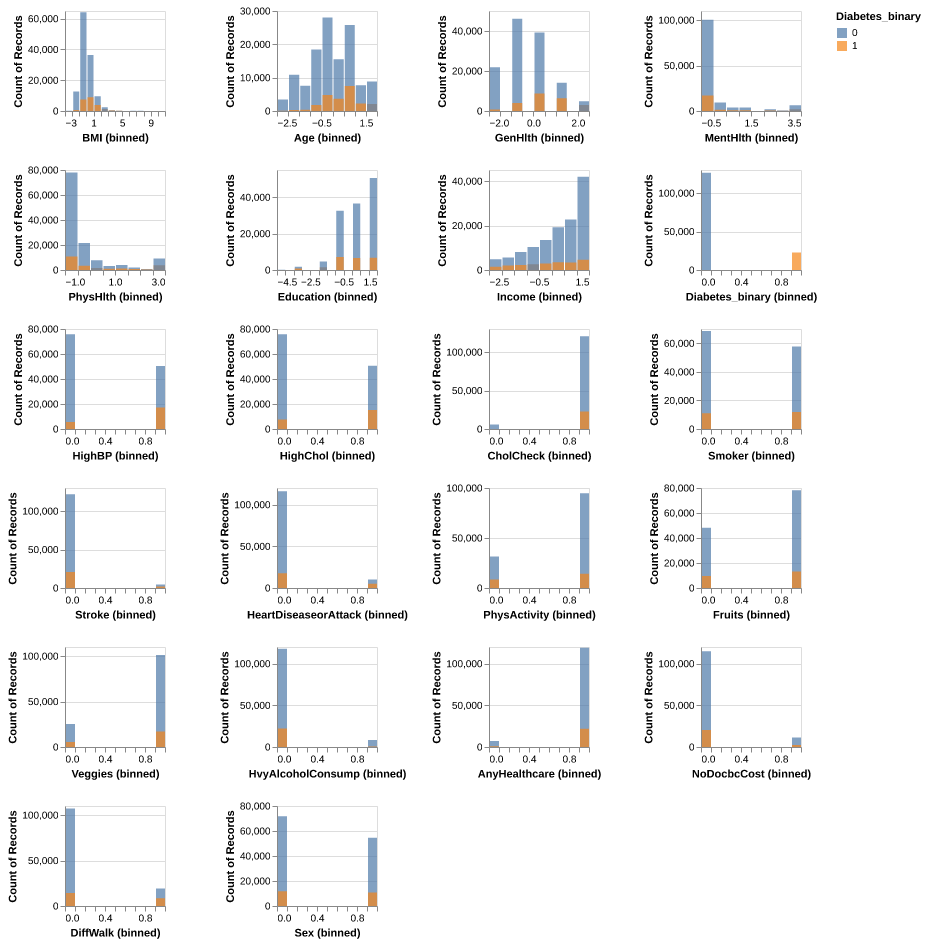
Fig. 1 Histogram distribution for each feature in the training data for both non-diabetic (0 & blue) and diabetic (1 & orange) class.#
Using a random search, we performed hyperparameter optimization on a knn model and a decision tree model. For the knn model, we found that the optimal value for n_neighbors was 50. For the decision tree model, we found the optimal value for max_depth to be 5.
We then compared the two optimized models to a dummy classifier and a logistic regression model. The optimized decision tree model out-performed all others, with a cross validation score of 0.852 (+/- 0.001). As seen in the table below (Figure 2), all of the scores were quite high for all models, even the dummy model. This is likely due to the imbalance in the target features, which should be addressed if this model were to be improved and used.
| fit_time | score_time | test_score | train_score | |
|---|---|---|---|---|
| Dummy | 0.034 (+/- 0.002) | 0.005 (+/- 0.001) | 0.847 (+/- 0.000) | 0.847 (+/- 0.000) |
| Decision tree | 0.309 (+/- 0.009) | 0.007 (+/- 0.000) | 0.852 (+/- 0.001) | 0.852 (+/- 0.000) |
| Logistic regression | 0.115 (+/- 0.005) | 0.006 (+/- 0.001) | 0.847 (+/- 0.000) | 0.847 (+/- 0.000) |
| Knn | 0.047 (+/- 0.002) | 1.222 (+/- 0.038) | 0.847 (+/- 0.000) | 0.847 (+/- 0.000) |
Fig. 2 Cross validation results for all optimized models, as a result of a random search hyperparamater optimization, compared to a dummy classifier.#
Although the logistic regression model did not have the best performance compared to the other models, with a cross-validation score of 0.847 (+/- 0.000) its coefficients were still useful in investigating which features were weighted as more important in indicating whether a person has diabetes/prediabetes. The results, shown in the table below (Figure 3), indicate that general health (GenHlth), is the largest predictor of diabetes/prediabetes. For this feature, patients were asked to rate their general health on a scale of 1-5 where 1 = excellent, 2 = very good, 3 = good, 4 = fair, 5 = poor. The next largest predictors of diabetes/prediabetes were body mass index (BMI), high blood pressure (HighBP), and age, respectively. All of these factors, unsurprisingly had a positive correlation with the presence of diabetes/prediabetes. It is important to note that this analysis does not allow us to make any causal inference. However, these results allows us to see which features are more strongly correlated with diabetes which could inform future studies or analyses.
| features | coefficients | |
|---|---|---|
| 14 | GenHlth | 0.55 |
| 4 | BMI | 0.40 |
| 19 | Age | 0.38 |
| 1 | HighBP | 0.37 |
| 2 | HighChol | 0.27 |
| 3 | CholCheck | 0.25 |
| 11 | HvyAlcoholConsump | -0.19 |
| 18 | Sex | 0.14 |
| 21 | Income | -0.09 |
| 16 | PhysHlth | -0.07 |
| 7 | HeartDiseaseorAttack | 0.07 |
| 17 | DiffWalk | 0.06 |
| 15 | MentHlth | -0.03 |
| 6 | Stroke | 0.02 |
| 9 | Fruits | -0.02 |
| 12 | AnyHealthcare | 0.02 |
| 20 | Education | -0.02 |
| 5 | Smoker | -0.01 |
| 8 | PhysActivity | -0.01 |
| 10 | Veggies | -0.01 |
| 13 | NoDocbcCost | 0.00 |
Fig. 3 Logistic regression coefficients by feature, ordered by absolute value of coefficient.#
To enhance the future iterations of this model for potential use in medical pre-diagnosis, we propose the following recommendations:
Begin by closely examining instances of 11179 misclassifications as well as 784, comparing them to correctly classified observations from both classes. The objective is to identify features driving misclassifications and explore potential feature engineering to improve the model’s accuracy on these problematic observations.
Investigate alternative classifiers to determine if enhanced predictions can be attained. A potential option is to employ a random forest classifier, which inherently considers feature interactions, in contrast to the k-NN model.
Acknowledge the significant class imbalance between diabetic and non-diabetic classes and address it in the preprocessor for subsequent analyses.
Enhance the model’s usability in diagnosis by providing and reporting probability estimates for predictions. This approach ensures that even if misclassifications persist, clinicians with domain knowledge can gauge the model’s confidence in its predictions.
References#
- BHG+17
Nilka Rios Burrows, Israel Hora, Linda S Geiss, Edward W Gregg, and Ann Albright. Incidence of end-stage renal disease attributed to diabetes among persons with diagnosed diabetes—united states and puerto rico, 2000–2014. Morbidity and Mortality Weekly Report, 66(43):1165, 2017. URL: https://pubmed.ncbi.nlm.nih.gov/29095800/, doi:10.15585/mmwr.mm6643a2.
- PVG+11
Fabian Pedregosa, Gaël Varoquaux, Alexandre Gramfort, Vincent Michel, Bertrand Thirion, Olivier Grisel, Mathieu Blondel, Peter Prettenhofer, Ron Weiss, Vincent Dubourg, and others. Scikit-learn: machine learning in python. the Journal of machine Learning research, 12:2825–2830, 2011. doi:10.5555/1953048.2078195.
- SB23(1,2,3)
A. Sapra and P. Bhandari. Diabetes. 2023. [Updated 2023 Jun 21]. In: StatPearls [Internet]. Treasure Island (FL): StatPearls Publishing; 2023 Jan-. URL: https://www.ncbi.nlm.nih.gov/books/NBK551501/.
- VGH+18
Jacob VanderPlas, Brian Granger, Jeffrey Heer, Dominik Moritz, Kanit Wongsuphasawat, Arvind Satyanarayan, Eitan Lees, Ilia Timofeev, Ben Welsh, and Scott Sievert. Altair: interactive statistical visualizations for python. Journal of open source software, 3(32):1057, 2018. URL: https://joss.theoj.org/papers/10.21105/joss.01057, doi:10.21105/joss.01057.
- WesMcKinney10
Wes McKinney. Data Structures for Statistical Computing in Python. In Stéfan van der Walt and Jarrod Millman, editors, Proceedings of the 9th Python in Science Conference, 56 – 61. 2010. doi:10.25080/Majora-92bf1922-00a.