prepropyr-vignette
Chun Chieh(Jason) Chang, Bruhat Musunuru, Pan Fan
prepropyr-vignette.Rmd
library(prepropyr)Introduction
The functions in prepropyr were developed to perform a quick EDA, data imputation and data scaling. Currently prepropyr contains three independent functions, this vignette outlines how to use these functions with toy data.
eda()
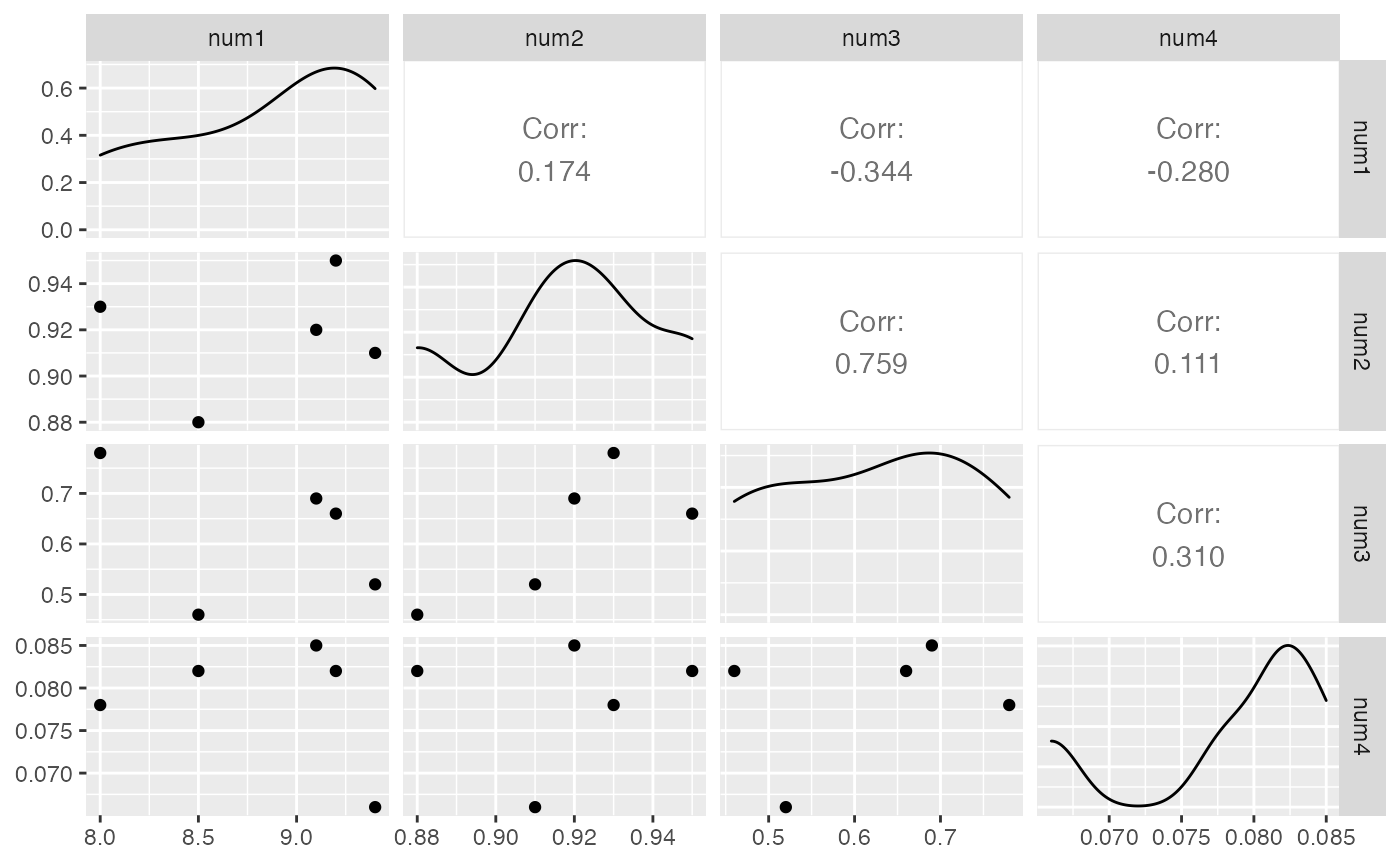
The eda() function helps to quickly explore the data by showing a pairplot and some summary statistics for a given dataframe .
df <- data.frame(num1 = c(8.5, 8, 9.2, 9.1, 9.4),
num2 = c(0.88, 0.93, 0.95 , 0.92 , 0.91),
num3 = c(0.46, 0.78, 0.66, 0.69, 0.52),
num4 = c(0.082, 0.078, 0.082, 0.085, 0.066),
cat1 = c("Good","Okay","Excellent","Terrible","Good"),
target = c(2,2,3,1,3))After calling the eda() function, we can get following outputs,please see the docstring for more available outputs.
result <- eda(df,"target")
#> Registered S3 method overwritten by 'GGally':
#> method from
#> +.gg ggplot2
result$nb_num_features
#> [1] 4
result$pairplot
imputation()
The imputation() function will impute missing data in a tibble/dataframe given the chosen method(mean, median)
test_df <- data.frame('a' = c(1,NA,3), 'b' = c(5,6,NA), 'c' = c(NA,1,10))
test_df_imputed <- imputation(test_df, test_df, 'mean')
test_df_imputed
#> a b c
#> 1 1 5.0 5.5
#> 2 2 6.0 1.0
#> 3 3 5.5 10.0
scaler()
This function scales numerical features based on scaling requirement(standardization, minmax Scaling) in a data.frame
X_train <- data.frame('a' = c(1,2,3), 'b' = c(5,6,3), 'c' = c(2,1,10))
X_test <- data.frame('a' = c(1,5,3), 'b' = c(5,6,5), 'c' = c(2,5,10))
X_Valid <- data.frame('a' = c(5,5,3), 'b' = c(5,6,5), 'c' = c(2,5,10))
scaled_df <- scaler(X_train, X_Valid, X_test, scaler_type='standardization')
scaled_df
#> $train
#> a b c
#> 1 -1 0.2182179 -0.4730162
#> 2 0 0.8728716 -0.6757374
#> 3 1 -1.0910895 1.1487535
#>
#> $test
#> a b c
#> 1 -1 0.2182179 -0.4730162
#> 2 3 0.8728716 0.1351475
#> 3 1 0.2182179 1.1487535
#>
#> $valid
#> a b c
#> 1 3 0.2182179 -0.4730162
#> 2 3 0.8728716 0.1351475
#> 3 1 0.2182179 1.1487535