
Lecture 6: L2- and L1-Regularization#
UBC Master of Data Science program, 2022-23
Instructor: Varada Kolhatkar
Atrribution: A lot of this material is adapted from CPSC 340 lecture notes.
Imports#
import os
import sys
sys.path.append("code/.")
%matplotlib inline
import matplotlib.pyplot as plt
import mglearn
import numpy as np
import numpy.linalg as npla
import numpy.random as npr
import pandas as pd
from plotting_functions import *
from sklearn import datasets
from sklearn.dummy import DummyClassifier, DummyRegressor
from sklearn.impute import SimpleImputer
from sklearn.linear_model import LinearRegression, LogisticRegression, Ridge, RidgeCV
from sklearn.model_selection import cross_val_score, cross_validate, train_test_split
from sklearn.preprocessing import PolynomialFeatures, StandardScaler
pd.set_option("display.max_colwidth", 200)
Learning outcomes#
From this lecture, students are expected to be able to:
Broadly explain L2 regularization (Ridge).
Use L2 regularization (Ridge) using
sklearn.Compare L0- and L2-regularization.
Explain the relation between the size of regression weights, overfitting, and complexity hyperparameters.
Explain the effect of training data size and effect of regularization.
Explain the general idea of L1-regularization.
Learn to be skeptical about interpretation of the coefficients.
Use L1-regularization (Lasso) using
sklearn.Discuss sparsity in L1-regularization.
Compare L0-, L1-, and L2-regularization.
Use L1 regularization for feature selection.
Explain the importance of scaling when using L1- and L2-regularization
Broadly explain how L1 and L2 regularization behave in the presence of collinear features.
Recap#
Idea of regularization: Pick the line/hyperplane with smaller slope#
Assuming the models below (dashed and solid green lines) have the same training error and if you are forced to choose one of them, which one would you pick?
Pick the solid green line because its slope is smaller.
Small change in \(x_i\) has a smaller change in prediction \(y_i\)
Since \(w\) is less sensitive to the training data, it’s likely to generalize better.
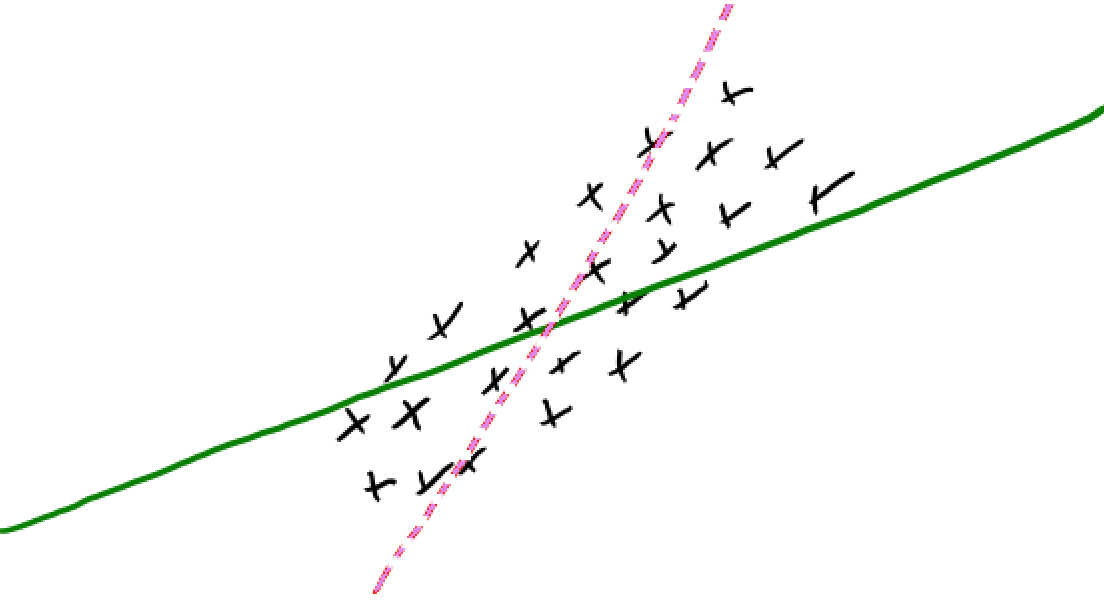
Regularization is a technique for controlling model complexity.
Instead of minimizing just the Loss, minimize Loss + \(\lambda \) Model complexity
Loss: measures how well the model fits the data.
Model complexity: measures model complexity (also referred to as regularization term)
A scalar \(\lambda\) decides the overall impact of the regularization term.
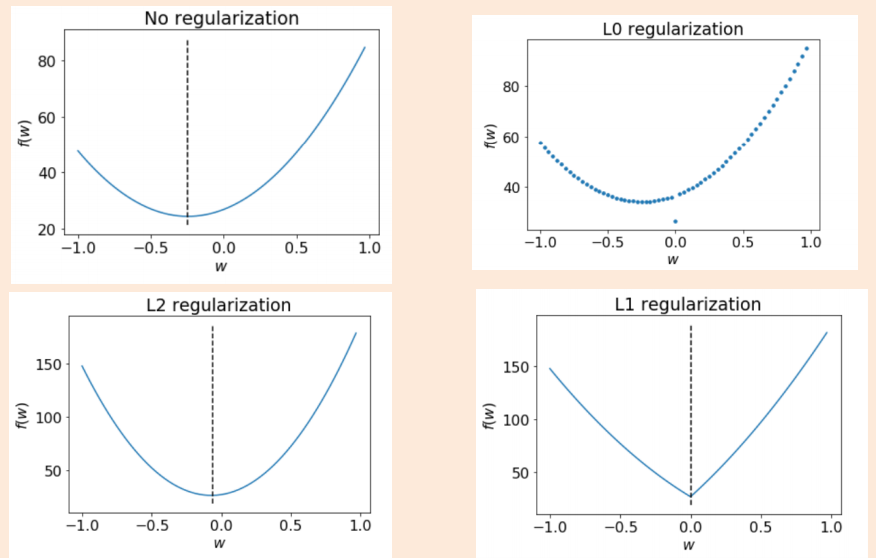
How to quantify model complexity?#
Total number of features with non-zero weights
L0 regularization: quantify model complexity as \(\lVert w \rVert_0\), L0 norm of \(w\).
As a function of weights: A feature weight with high absolute value is more complex than the feature weight with low absolute value.
L2 regularization: quantify model complexity as \(\lVert w \rVert_2^2\), square of the L2 norm of \(w\).
L1 regularization: quantify model complexity as \(\lVert w \rVert_1\), L1 norm of \(w\).
L0-regularization#
L0-regularization.
Regularization parameter \(\lambda\) controls the strength of regularization.
To increase the degrees of freedom by one, need to decrease the error by \(\lambda\).
Prefer smaller degrees of freedom if errors are similar.
Can’t optimize because the function is discontinuous in \(\lVert w\rVert_0\)
Search over possible models
L2-regularization#
Standard regularization strategy is L2-regularization.
Also referred to as Ridge.
The loss function has an L2 penalty term.
Regularization parameter \(\lambda\) controls the strength of regularization.
bigger \(\lambda\) \(\rightarrow\) more regularization \(\rightarrow\) lower weights
Setting \(\lambda = 0\) is the same as using OLS
Low training error, high variance, bigger weights, likely to overfit
Setting \(\lambda\) to a large value leads to smaller weights
Higher training error, low variance, high bias, smaller weights, likely to underfit
We want to strike a balance between these two extremes, which we can control by optimizing the value for \(\lambda\).
sklearn’s Ridge or L2 regularization#
Let’s bring back sklearn's California housing dataset.
from sklearn.datasets import fetch_california_housing
from sklearn.preprocessing import PolynomialFeatures
housing = fetch_california_housing(as_frame=True)
X = housing["data"]
y = housing["target"]
pf = PolynomialFeatures(degree=2, include_bias=False)
X = pf.fit_transform(X)
X_train_poly, X_test_poly, y_train, y_test = train_test_split(X, y, random_state=0)
ss = StandardScaler()
X_train = ss.fit_transform(X_train_poly)
X_test = ss.transform(X_test_poly)
X_train.shape
(15480, 44)
Let’s explore L2 regularization with this dataset.
Example: L2-Regularization “Shrinking”#
\(\lambda\) in the formulation above is
alphainsklearn.We get least squares with \(\lambda = 0\).
As we increase \(\alpha\)
\(\lVert Xw - y\rVert_2^2\) goes \(\uparrow\)
\(\lVert w\rVert_2^2\) goes \(\downarrow\)
Though individual \(w_j\) can increase or decrease with lambda because we use the L2-norm, the large ones decrease the most.
def lr_loss_squared(w, X, y): # define OLS loss function
return np.sum((y - X @ w) ** 2)
from numpy.linalg import norm
alphas = [0, 0.01, 0.1, 1.0, 10, 100, 1000, 1000000]
weight_shrinkage_data = {'alpha':[], '||Xw - y||^2':[], '||w||^2':[], 'intercept':[], 'train score':[], 'test score':[]}
for (i, alpha) in enumerate(alphas):
r = Ridge(alpha=alpha)
r.fit(X_train, y_train)
weight_shrinkage_data['alpha'].append(alpha)
OLS = np.round(lr_loss_squared(r.coef_, X_train, y_train), 4)
l2_term = np.round(norm(r.coef_, 2)**2, 4)
intercept = np.round(r.intercept_,4)
weight_shrinkage_data['||Xw - y||^2'].append(OLS)
weight_shrinkage_data['||w||^2'].append(l2_term)
weight_shrinkage_data['intercept'].append(intercept)
weight_shrinkage_data['train score'].append(np.round(r.score(X_train, y_train), 2))
weight_shrinkage_data['test score'].append(np.round(r.score(X_test, y_test), 2))
pd.DataFrame(weight_shrinkage_data)
| alpha | ||Xw - y||^2 | ||w||^2 | intercept | train score | test score | |
|---|---|---|---|---|---|---|
| 0 | 0.00 | 73099.4882 | 7634.4579 | 2.0744 | 0.69 | -0.73 |
| 1 | 0.01 | 73113.1963 | 4239.9359 | 2.0744 | 0.69 | -0.17 |
| 2 | 0.10 | 73222.8589 | 888.3987 | 2.0744 | 0.68 | 0.62 |
| 3 | 1.00 | 73437.3112 | 77.0152 | 2.0744 | 0.67 | 0.02 |
| 4 | 10.00 | 73597.9629 | 8.4983 | 2.0744 | 0.66 | -3.80 |
| 5 | 100.00 | 73777.7373 | 2.5195 | 2.0744 | 0.65 | -3.37 |
| 6 | 1000.00 | 74363.2804 | 0.7118 | 2.0744 | 0.62 | 0.10 |
| 7 | 1000000.00 | 85565.9420 | 0.0008 | 2.0744 | 0.08 | 0.08 |
In this example, as alpha goes up
\(\lVert Xw - y\rVert_2^2\) is increasing
\(\lVert w\rVert_2^2\) is decreasing; weight become smaller and smaller
The intercept stays the same. It’s the mean of the target – dummy model prediction.
We are getting best test score for
alpha=0.10.The total loss is worse than OLS but it’s helping with generalization.
The weights become smaller but do not become zero#
As
alphagoes up, we increase the regularization strength.The weights become smaller and smaller but tend not to become exactly zero.
alphas = [0, 0.01, 1, 1000]
plt.figure(figsize=(6, 4))
markers = ['o','*', 'v','^']
ridge_models = {}
for (i, alpha) in enumerate(alphas):
r = Ridge(alpha = alpha)
r.fit(X_train, y_train)
plt.plot(r.coef_, markers[i], markersize=5, label="Ridge alpha="+ str(alpha))
plt.xlabel("Coefficient index")
plt.ylabel("Coefficient magnitude")
plt.hlines(0, 0, len(r.coef_))
plt.ylim(-25, 25)
plt.legend(loc="best",fontsize=8);
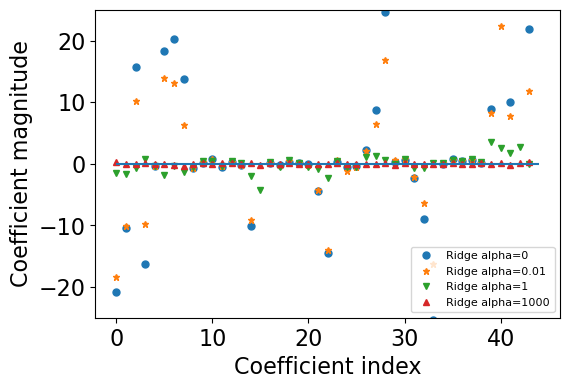
Let’s try a very large alpha value, which is likely to make weights very small.
ridge10000 = Ridge(alpha=10000).fit(X_train, y_train)
print("Total number of features", X_train.shape[1])
print("Number of non-zero weights: ", np.sum(ridge10000.coef_ != 0))
Total number of features 44
Number of non-zero weights: 44
All features have non-zero weights.
Regularization path#
Let’s trace the journey of coefficients of a few features with different values of alpha.
feature_names = pf.get_feature_names_out()
Let’s examine the coefficients
r10 = Ridge(alpha=0.10)
r10.fit(X_train, y_train)
coefs = (
pd.DataFrame(r10.coef_, feature_names, columns=["coef"])
.query("coef != 0")
.sort_values("coef")
.reset_index()
.rename(columns={"index": "variable"})
)
coefs.style.background_gradient('PuOr')
| variable | coef | |
|---|---|---|
| 0 | MedInc Longitude | -16.762873 |
| 1 | MedInc | -10.178537 |
| 2 | HouseAge Longitude | -9.608717 |
| 3 | HouseAge | -6.986869 |
| 4 | MedInc Latitude | -5.683734 |
| 5 | HouseAge Latitude | -2.931366 |
| 6 | AveBedrms Longitude | -2.152434 |
| 7 | AveBedrms Latitude | -1.934514 |
| 8 | Longitude | -1.637599 |
| 9 | AveRooms AveBedrms | -1.571689 |
| 10 | AveBedrms AveOccup | -1.536152 |
| 11 | MedInc^2 | -0.540772 |
| 12 | AveRooms Population | -0.518624 |
| 13 | HouseAge AveOccup | -0.414707 |
| 14 | HouseAge AveRooms | -0.376728 |
| 15 | MedInc AveBedrms | -0.280714 |
| 16 | Population | -0.106295 |
| 17 | MedInc AveOccup | -0.072894 |
| 18 | AveBedrms | -0.067099 |
| 19 | Population^2 | 0.050877 |
| 20 | HouseAge Population | 0.076735 |
| 21 | AveOccup^2 | 0.177787 |
| 22 | HouseAge^2 | 0.197444 |
| 23 | MedInc HouseAge | 0.292222 |
| 24 | MedInc Population | 0.358598 |
| 25 | HouseAge AveBedrms | 0.495107 |
| 26 | MedInc AveRooms | 0.544633 |
| 27 | Population Latitude | 0.573911 |
| 28 | Population AveOccup | 0.603423 |
| 29 | AveBedrms^2 | 0.620403 |
| 30 | AveBedrms Population | 0.780217 |
| 31 | AveRooms | 0.908226 |
| 32 | AveRooms^2 | 0.940577 |
| 33 | Longitude^2 | 0.944154 |
| 34 | Population Longitude | 1.199605 |
| 35 | AveRooms AveOccup | 1.697423 |
| 36 | AveOccup | 2.478821 |
| 37 | AveRooms Latitude | 2.523284 |
| 38 | AveRooms Longitude | 3.693662 |
| 39 | Latitude | 3.830685 |
| 40 | Latitude^2 | 4.330258 |
| 41 | AveOccup Latitude | 6.270548 |
| 42 | AveOccup Longitude | 9.143782 |
| 43 | Latitude Longitude | 10.627639 |
feats = ['MedInc', 'AveBedrms', 'Population', 'MedInc Longitude', 'Population Longitude', 'Latitude Longitude']
msk = [feat in feats for feat in feature_names]
#feature_names
r10.coef_[msk]
array([-10.17853657, -0.06709887, -0.10629543, -16.7628735 ,
1.19960525, 10.6276393 ])
# Attribution: Code adapted from here: https://scikit-learn.org/stable/auto_examples/linear_model/plot_ridge_path.html
import numpy as np
import matplotlib.pyplot as plt
from sklearn import linear_model
n_alphas = 200
alphas = np.logspace(-3, 3, n_alphas)
coefs = []
for a in alphas:
ridge = linear_model.Ridge(alpha=a, fit_intercept=False)
ridge.fit(X_train, y_train)
coefs.append(ridge.coef_[msk])
ax = plt.gca()
ax.plot(alphas, coefs)
ax.set_xscale("log")
#ax.set_xlim(ax.get_xlim()[::-1]) # reverse axis
plt.xlabel("alpha")
plt.ylabel("weights")
plt.title("Ridge coefficients as a function of the regularization")
plt.axis("tight")
ax.legend(feats, loc="best", fontsize=11)
plt.show()
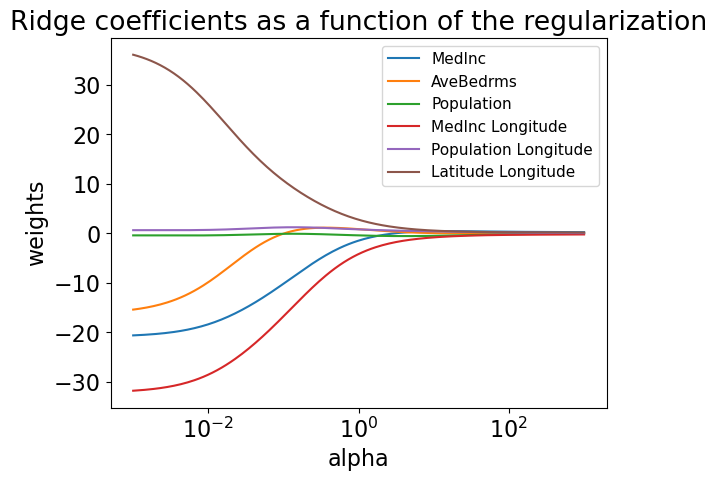
The coefficients are very close to zero but not exactly zero. That said, they won’t have much impact on prediction.
Regularization learning curves#
plot_learning_curve(Ridge(alpha=0.01), X_train, y_train)
plot_learning_curve(Ridge(alpha=10.0), X_train, y_train)
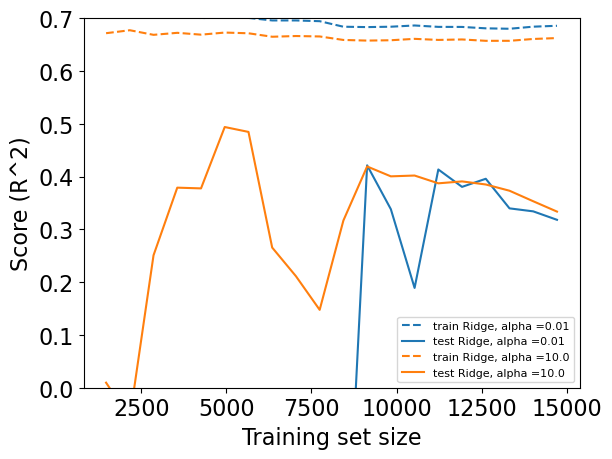
As one would expect, the training score is higher than the test score for all dataset sizes
Training score of
Ridgewithalpha=0.01> Training score ofRidgewithalpha=10(more regularization)However, the test score of
Ridgewithalpha=10> Test score ofRidgewithalpha=0.01for smaller training sizes.As more and more data becomes available to the model, both models improve. Ridge with less regularization catches up with ridge with more regularization and actually performs better.
How to pick \(\lambda\)?#
Theory: as \(n\) grows \(\lambda\) should be in the range \(O(1)\) to \(\sqrt{n}\).
Practice: optimize validation set or cross-validation error.
Almost always decreases the test error.
(Optional) Should we regularize the y-intercept?#
No!
Why encourage it to be closer to zero? (It could be anywhere.)
You should be allowed to shift function up/down globally.
Yes!
Useful for optimization; It makes the solution unique and it easier to compute \(w\)
Compromise: regularize by a smaller amount than other variables. $\(f(w) = \lVert Xw + w_0 - y\rVert^2 + \frac{\lambda_1}{2}\lVert w\rVert^2 + \frac{\lambda_2}{2}w_0^2\)$
Some properties of L2-regularization#
Solution \(w\) is unique. It has to do with the smoothness of the function.
We are not going into mathematical details. If interested see slide 20 from CPSC 340.
Almost always improves the validation error.
No collinearity issues.
Less sensitive to changes in \(X\).
Gradient descent (optimization algorithm) converges faster (bigger \(\lambda\) means fewer iterations). (You’ll learn about Grafient descent in 572.)
Worst case: just set \(\lambda\) small and get the same performance
Summary: L2-regularization#
Change the loss function by adding a continuous L2-penalty on the model complexity.
Best parameter \(\lambda\) almost already leads to improved validation error.
L2-regularized least squares is also called “ridge regression”.
Can be solved as a linear system like least squares.
Some benefits of L2 regularization
Solution is unique.
Less sensitive to data.
Fast.
❓❓ Questions for you (recap)#
iClicker Exercise 6.1#
iClicker cloud join link: https://join.iclicker.com/C0P55
Select all of the following statements which are TRUE.
(A) Introducing L2 regularization to the model means making it less sensitive to changes in \(X\).
(B) Introducing L2 regularization to the model can results in worse performance on the training set.
(C) L2 regularization shrinks the weights but all \(w_j\)s tend to be non-zero.
V’s answers: A, B, C
Break (~5 mins)#

L1 regularization#
An alternative to
Ridge(L2-regularization for least squares) isLasso.Instead of L0- or L2-norm, regularize with L1-norm.
\(\lambda \rightarrow\) regularization strength
\(\lVert w\rVert_1 \rightarrow\) L1-norm of \(w\)
Objective balances getting low error vs. having small values for \(w_j\)
Similarities with L2-regularization#
L1-regularization $\(f(w) = \lVert Xw - y\rVert_2^2 + \lambda \lVert w\rVert_1\)$
L2-regularization $\(f(w) = \lVert Xw - y\rVert_2^2 + \lambda \lVert w\rVert_2^2\)$
Both shrink weights.
Both result in lower validation error.
Do not confuse the L1 loss (absolute loss) with L1-regularization!
Terminology and notation: Ridge and Lasso#
Linear regression model that uses L2 regularization is called Ridge or Tikhonov regularization.
Linear regression model that uses L1 regularization is called Lasso.
class sklearn.linear_model.Lasso(alpha=1.0, fit_intercept=True, normalize=False, precompute=False, copy_X=True, max_iter=1000, tol=0.0001, warm_start=False, positive=False, random_state=None, selection=’cyclic’)
L1-regularization#
The consequence of using L1-norm is that some features are exactly zero, which means that the features are entirely ignored by the model.
This can be considered as a form of feature selection!!
L1-regularization simultaneously regularizes and selects features.
Very fast alternative to search and score methods
Let’s apply Lasso on the California housing data.
X_train.shape
(15480, 44)
from sklearn.linear_model import Lasso
lasso = Lasso().fit(X_train, y_train)
print("Training set score: {:.2f}".format(lasso.score(X_train, y_train)))
print("Test set score: {:.2f}".format(lasso.score(X_test, y_test)))
print("Number of features used:", np.sum(lasso.coef_ != 0))
Training set score: 0.00
Test set score: -0.00
Number of features used: 0
That’s strange. It’s not using any features – like dummy model. It’s underfitting.
Similar to
Ridgeit also has a regularization hyperparameteralpha.Let’s decrease it to reduce underfitting.
Let’s examine weight shrinkage for l1 regularization
Lasso loss is not a smooth function.
Slower compared to Ridge.
from numpy.linalg import norm
alphas = [0, 0.0001, 0.01, 0.1, 1]
weight_shrinkage_data = {'alpha':[], '||Xw - y||^2':[], '||w||_1':[], 'intercept':[], 'train score':[], 'test score':[]}
for (i, alpha) in enumerate(alphas):
print('alpha: ', alpha)
l = Lasso(alpha=alpha, max_iter=10000)
l.fit(X_train, y_train)
weight_shrinkage_data['alpha'].append(alpha)
OLS = np.round(lr_loss_squared(l.coef_, X_train, y_train), 4)
l1_term = np.round(norm(r.coef_, 1), 4)
intercept = np.round(l.intercept_, 4)
weight_shrinkage_data['||Xw - y||^2'].append(OLS)
weight_shrinkage_data['||w||_1'].append(l1_term)
weight_shrinkage_data['intercept'].append(intercept)
weight_shrinkage_data['train score'].append(np.round(l.score(X_train, y_train), 2))
weight_shrinkage_data['test score'].append(np.round(l.score(X_test, y_test), 2))
alpha: 0
/var/folders/b3/g26r0dcx4b35vf3nk31216hc0000gr/T/ipykernel_27748/2689048393.py:8: UserWarning: With alpha=0, this algorithm does not converge well. You are advised to use the LinearRegression estimator
l.fit(X_train, y_train)
/Users/kvarada/opt/miniconda3/envs/573/lib/python3.10/site-packages/sklearn/linear_model/_coordinate_descent.py:648: UserWarning: Coordinate descent with no regularization may lead to unexpected results and is discouraged.
model = cd_fast.enet_coordinate_descent(
/Users/kvarada/opt/miniconda3/envs/573/lib/python3.10/site-packages/sklearn/linear_model/_coordinate_descent.py:648: ConvergenceWarning: Objective did not converge. You might want to increase the number of iterations, check the scale of the features or consider increasing regularisation. Duality gap: 3.394e+03, tolerance: 2.066e+00 Linear regression models with null weight for the l1 regularization term are more efficiently fitted using one of the solvers implemented in sklearn.linear_model.Ridge/RidgeCV instead.
model = cd_fast.enet_coordinate_descent(
alpha: 0.0001
/Users/kvarada/opt/miniconda3/envs/573/lib/python3.10/site-packages/sklearn/linear_model/_coordinate_descent.py:648: ConvergenceWarning: Objective did not converge. You might want to increase the number of iterations, check the scale of the features or consider increasing regularisation. Duality gap: 1.778e+03, tolerance: 2.066e+00
model = cd_fast.enet_coordinate_descent(
alpha: 0.01
alpha: 0.1
alpha: 1
pd.DataFrame(weight_shrinkage_data)
| alpha | ||Xw - y||^2 | ||w||_1 | intercept | train score | test score | |
|---|---|---|---|---|---|---|
| 0 | 0.0000 | 73398.6742 | 4.3674 | 2.0744 | 0.67 | 0.52 |
| 1 | 0.0001 | 73462.0080 | 4.3674 | 2.0744 | 0.67 | 0.20 |
| 2 | 0.0100 | 74463.3147 | 4.3674 | 2.0744 | 0.62 | 0.33 |
| 3 | 0.1000 | 76723.2012 | 4.3674 | 2.0744 | 0.51 | 0.48 |
| 4 | 1.0000 | 87271.1329 | 4.3674 | 2.0744 | 0.00 | -0.00 |
r = Ridge(alpha = 0.1).fit(X_train, y_train)
print("Training set score: {:.2f}".format(r.score(X_train, y_train)))
print("Test set score: {:.2f}".format(r.score(X_test, y_test)))
print("Number of features used:", np.sum(r.coef_ != 0))
Training set score: 0.68
Test set score: 0.62
Number of features used: 44
lasso = Lasso(alpha=0.1, max_iter=10000).fit(X_train, y_train)
print("Training set score: {:.2f}".format(lasso.score(X_train, y_train)))
print("Test set score: {:.2f}".format(lasso.score(X_test, y_test)))
print("Number of features used:", np.sum(lasso.coef_ != 0))
Training set score: 0.51
Test set score: 0.48
Number of features used: 3
In this case we are not really getting better results with Lasso.
In fact, Lasso is not able to do better than linear regression.
Often you might see better scores than
Ridgewith fewer features.If
alphais too low, we reduce the effect of overfitting and the results are similar to linear regression.
The weights become smaller and eventually become zero.#
Let’s plot coefficients of Lasso.
alphas = [0, 0.01, 0.1, 1]
plt.figure(figsize=(6, 4))
markers = ['o','*', 'v','^']
lasso_models = {}
for (i, alpha) in enumerate(alphas):
lasso = Lasso(alpha = alpha, max_iter=10000)
lasso.fit(X_train, y_train)
plt.plot(r.coef_, markers[i], markersize=5, label="Lasso alpha="+ str(alpha))
plt.xlabel("Coefficient index")
plt.ylabel("Coefficient magnitude")
plt.hlines(0, 0, len(lasso.coef_))
plt.ylim(-25, 25)
plt.legend(loc="best",fontsize=8);
/var/folders/b3/g26r0dcx4b35vf3nk31216hc0000gr/T/ipykernel_27748/562400412.py:6: UserWarning: With alpha=0, this algorithm does not converge well. You are advised to use the LinearRegression estimator
lasso.fit(X_train, y_train)
/Users/kvarada/opt/miniconda3/envs/573/lib/python3.10/site-packages/sklearn/linear_model/_coordinate_descent.py:648: UserWarning: Coordinate descent with no regularization may lead to unexpected results and is discouraged.
model = cd_fast.enet_coordinate_descent(
/Users/kvarada/opt/miniconda3/envs/573/lib/python3.10/site-packages/sklearn/linear_model/_coordinate_descent.py:648: ConvergenceWarning: Objective did not converge. You might want to increase the number of iterations, check the scale of the features or consider increasing regularisation. Duality gap: 3.394e+03, tolerance: 2.066e+00 Linear regression models with null weight for the l1 regularization term are more efficiently fitted using one of the solvers implemented in sklearn.linear_model.Ridge/RidgeCV instead.
model = cd_fast.enet_coordinate_descent(
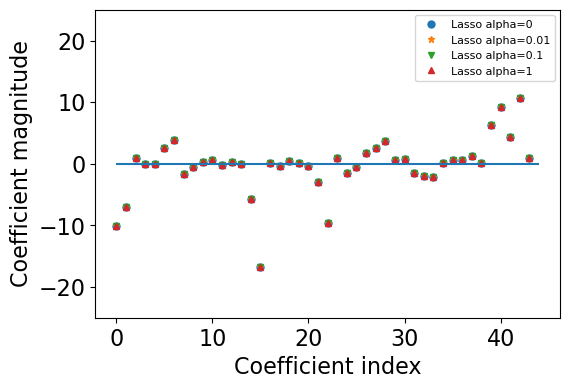
For
alpha = 100most of the coefficients are zero.With smaller values of
alphawe get less and less regularization.
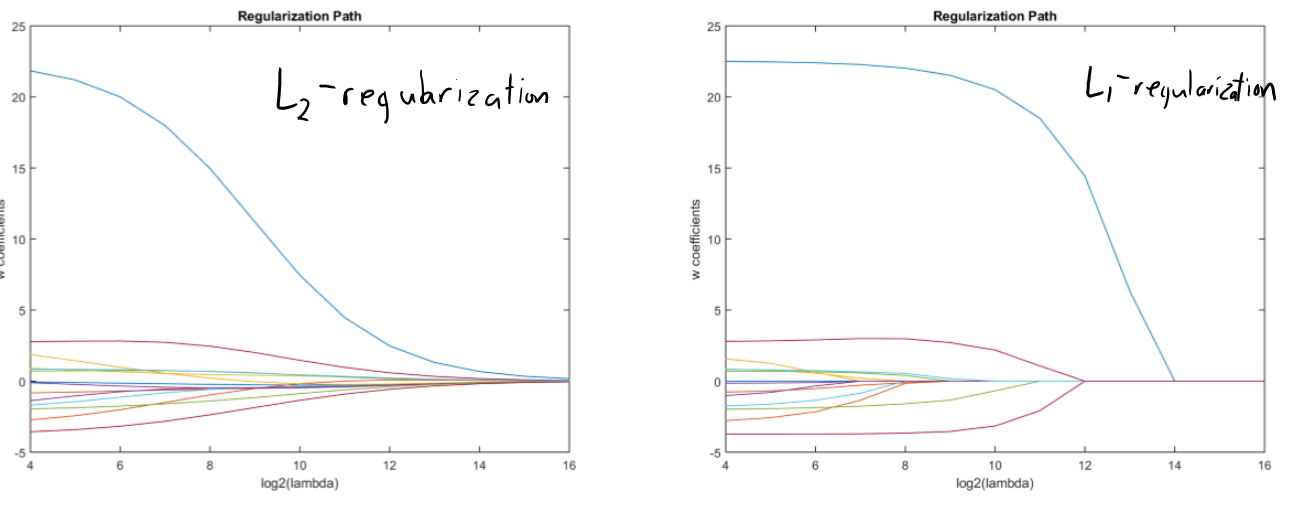
I’m not showing L1 regularization path on our data because it takes too long to run.
But you’re likely to see this type of plots in the context of regularization.
Terminology and notation: Sparsity#
We say a linear function is sparse if most of the coefficients are zero.
Example: Here only 2 out of 8 coefficients are non-zero and so it is a sparse function. $\(0x_1 + 0.45 x_2 + 0 x_3 + 0x_4 + 1.2x_5 + 0x_6 + 0x_7 + 0x_8\)$
L0- and L1-regularization encourage sparsity.
Example: L0 vs. L1 vs. L2#
Consider problem where 3 vectors can get minimum training error:
Without regularization, we could choose any of these 3.
We are assuming that all have the same error, so regularization will “break tie”.
Which one would you choose with each of L0, L1, L2 regularization?
Which one would you choose with L0 regularization?#
What are the L0 norms of different weight vectors?
\(\lVert w^1\rVert_0 = 2\)
\(\lVert w^2\rVert_0 = 1\)
\(\lVert w^3\rVert_0 = 2\)
L0 regularization would choose \(w^2\).
Which one would you choose with L1 regularization?#
What are the L1 norms of different weight vectors?
\(\lVert w^1\rVert_1 = 100.02\)
\(\lVert w^2\rVert_1 = 100\)
\(\lVert w^3\rVert_1 = 100.01\)
L1-regularization would pick \(w^2\)
L1-regularization focuses on decreasing all \(w_j\) until they are 0.
Which one would you choose with L2 regularization?#
What are the L2 norms of different weight vectors?
\(\lVert w^1\rVert_2^2 = (100)^2 + (0.02)^2 = 10000.0004\)
\(\lVert w^2\rVert_2^2 = (100)^2 = 10000\)
\(\lVert w^3\rVert_2^2 = (99.99)^2 + (0.02)^2 = 9998.0005\)
L2-regularization would pick \(w^3\)
L2-regularization focuses on decreasing largest \(w_j\) smaller
(Optional) Sparsity and Regularization#
Minimizing \(\lVert Xw - y\rVert_2^2 + \lambda \lVert w\rVert_2^2\) is equivalent to minimizing \(\lVert Xw - y\rVert_2^2\) subject to the constraint that \(\lVert w\rVert_2^2 \leq t(\delta)\)
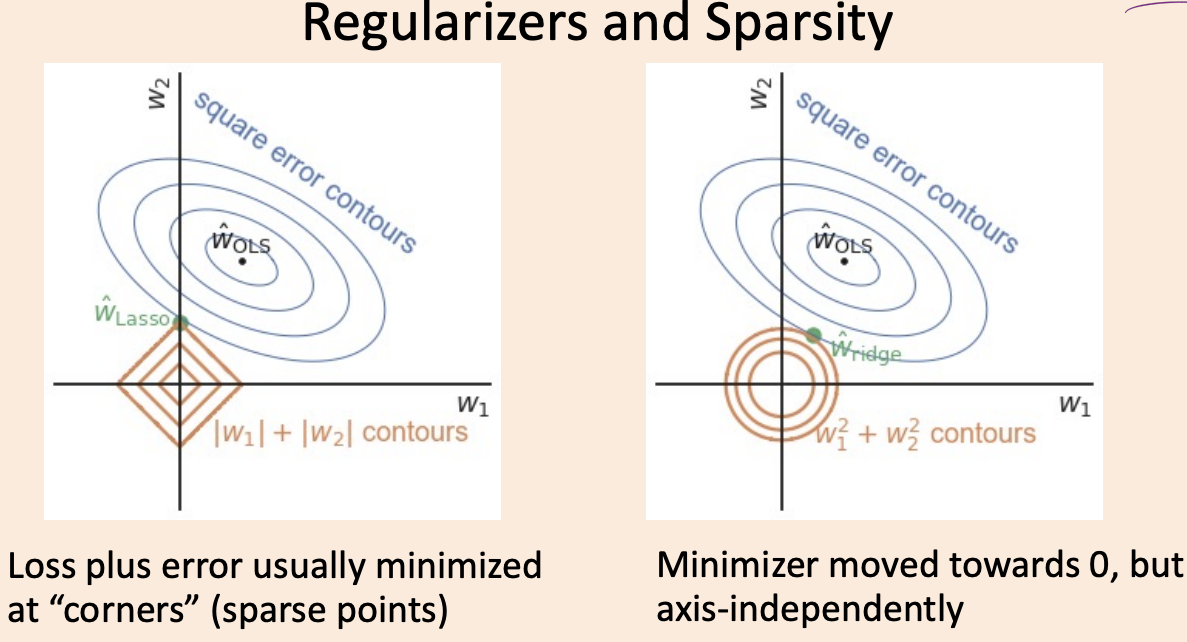

Sparsity and Regularization (with d=1)#
Some properties of L1 regularization#
Almost always improves the validation error.
Can learn with exponential number of irrelevant features.
Less sensitive to changes in \(X\).
The solution is not unique. (If interested in more explanation on this, see slide 43 in this slide deck.)
Feature selection using L1 regularization#
Feature selection methods we have seen so far:
RFE
Search and score with L0-regularization (e.g., forward search)
An effective way of feature selection: L1-regularization
from sklearn.ensemble import RandomForestRegressor
from sklearn.feature_selection import SelectFromModel
from sklearn.linear_model import Lasso, LassoCV
housing = fetch_california_housing(as_frame=True)
X = housing["data"]
y = housing["target"]
X_train, X_test, y_train, y_test = train_test_split(X, y, random_state=0)
pipe_l1_rf = make_pipeline(
StandardScaler(),
SelectFromModel(Lasso(alpha=0.01, max_iter=100000)),
RandomForestRegressor(),
)
X.shape
(20640, 8)
pipe_l1_rf.fit(X_train, y_train)
pipe_l1_rf.score(X_train, y_train)
0.9731789734697873
print("Training set score: {:.2f}".format(pipe_l1_rf.score(X_train, y_train)))
print("Test set score: {:.2f}".format(pipe_l1_rf.score(X_test, y_test)))
Training set score: 0.97
Test set score: 0.79
print(
"Number of features used:",
np.sum(pipe_l1_rf.named_steps["selectfrommodel"].estimator_.coef_ != 0),
)
Number of features used: 7
Regularized models for classification#
Regularized logistic regression#
Regularization is not limited to least squares.
We can add L1 and L2 penalty terms in other loss functions as well.
Let’s look at logistic regression with L1- and L2-regularization.
from sklearn.datasets import load_breast_cancer
breast_cancer = load_breast_cancer()
# print(breast_cancer.keys())
# print(breast_cancer.DESCR)
breast_cancer_df = pd.DataFrame(breast_cancer.data, columns=breast_cancer.feature_names)
breast_cancer_df["target"] = breast_cancer.target
train_df, test_df = train_test_split(breast_cancer_df, test_size=0.2, random_state=2)
X_train, y_train = train_df.drop(columns=["target"]), train_df["target"]
X_test, y_test = test_df.drop(columns=["target"]), test_df["target"]
X_train.head()
| mean radius | mean texture | mean perimeter | mean area | mean smoothness | mean compactness | mean concavity | mean concave points | mean symmetry | mean fractal dimension | ... | worst radius | worst texture | worst perimeter | worst area | worst smoothness | worst compactness | worst concavity | worst concave points | worst symmetry | worst fractal dimension | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 560 | 14.05 | 27.15 | 91.38 | 600.4 | 0.09929 | 0.11260 | 0.04462 | 0.04304 | 0.1537 | 0.06171 | ... | 15.30 | 33.17 | 100.20 | 706.7 | 0.1241 | 0.22640 | 0.1326 | 0.10480 | 0.2250 | 0.08321 |
| 428 | 11.13 | 16.62 | 70.47 | 381.1 | 0.08151 | 0.03834 | 0.01369 | 0.01370 | 0.1511 | 0.06148 | ... | 11.68 | 20.29 | 74.35 | 421.1 | 0.1030 | 0.06219 | 0.0458 | 0.04044 | 0.2383 | 0.07083 |
| 198 | 19.18 | 22.49 | 127.50 | 1148.0 | 0.08523 | 0.14280 | 0.11140 | 0.06772 | 0.1767 | 0.05529 | ... | 23.36 | 32.06 | 166.40 | 1688.0 | 0.1322 | 0.56010 | 0.3865 | 0.17080 | 0.3193 | 0.09221 |
| 203 | 13.81 | 23.75 | 91.56 | 597.8 | 0.13230 | 0.17680 | 0.15580 | 0.09176 | 0.2251 | 0.07421 | ... | 19.20 | 41.85 | 128.50 | 1153.0 | 0.2226 | 0.52090 | 0.4646 | 0.20130 | 0.4432 | 0.10860 |
| 41 | 10.95 | 21.35 | 71.90 | 371.1 | 0.12270 | 0.12180 | 0.10440 | 0.05669 | 0.1895 | 0.06870 | ... | 12.84 | 35.34 | 87.22 | 514.0 | 0.1909 | 0.26980 | 0.4023 | 0.14240 | 0.2964 | 0.09606 |
5 rows × 30 columns
y_train.value_counts(normalize=True)
1 0.632967
0 0.367033
Name: target, dtype: float64
from sklearn.metrics import f1_score, make_scorer, recall_score
custom_scorer = make_scorer(
f1_score, pos_label=0
) # note the syntax to change the positive label for f1 score
scoring_metric = custom_scorer
results_classification = {}
Let’s try DummyClassifier
dummy = DummyClassifier()
results_classification["dummy"] = mean_std_cross_val_scores(
dummy,
X_train,
y_train,
return_train_score=True,
scoring=scoring_metric,
)
pd.DataFrame(results_classification)
| dummy | |
|---|---|
| fit_time | 0.000 (+/- 0.001) |
| score_time | 0.000 (+/- 0.000) |
| test_score | 0.000 (+/- 0.000) |
| train_score | 0.000 (+/- 0.000) |
In
sklearn, by default logistic regression uses L2 regularization.The
Chyperparameter decides the strength of regularization.Unfortunately, interpretation of
Cis inverse oflambda.
pipe_lgr_l2 = make_pipeline(StandardScaler(), LogisticRegression())
results_classification["Logistic Regression L2"] = mean_std_cross_val_scores(
pipe_lgr_l2, X_train, y_train, return_train_score=True, scoring=scoring_metric
)
pd.DataFrame(results_classification)
| dummy | Logistic Regression L2 | |
|---|---|---|
| fit_time | 0.000 (+/- 0.001) | 0.005 (+/- 0.002) |
| score_time | 0.000 (+/- 0.000) | 0.001 (+/- 0.000) |
| test_score | 0.000 (+/- 0.000) | 0.970 (+/- 0.011) |
| train_score | 0.000 (+/- 0.000) | 0.985 (+/- 0.005) |
Weights are small but all of them are non-zero.
pipe_lgr_l2.fit(X_train, y_train)
l2_coefs = pipe_lgr_l2.named_steps["logisticregression"].coef_.flatten()
df = pd.DataFrame(l2_coefs, index=X_train.columns, columns=["l2_coefs"])
df
| l2_coefs | |
|---|---|
| mean radius | -0.539854 |
| mean texture | -0.252141 |
| mean perimeter | -0.494560 |
| mean area | -0.610451 |
| mean smoothness | -0.174066 |
| mean compactness | 0.577983 |
| mean concavity | -0.658777 |
| mean concave points | -0.980244 |
| mean symmetry | 0.119974 |
| mean fractal dimension | 0.406479 |
| radius error | -1.247956 |
| texture error | -0.103258 |
| perimeter error | -0.731201 |
| area error | -0.931584 |
| smoothness error | -0.230991 |
| compactness error | 0.614317 |
| concavity error | -0.032318 |
| concave points error | -0.206376 |
| symmetry error | 0.272883 |
| fractal dimension error | 0.739561 |
| worst radius | -1.040150 |
| worst texture | -1.096207 |
| worst perimeter | -0.885489 |
| worst area | -1.026863 |
| worst smoothness | -0.927290 |
| worst compactness | -0.075403 |
| worst concavity | -0.766568 |
| worst concave points | -0.732966 |
| worst symmetry | -0.669925 |
| worst fractal dimension | -0.520939 |
Let’s try logistic regression with L1 regularization.
Note that I’m using a different solver (optimizer) here because not all solvers support L1-regularization.
pipe_lgr_l1 = make_pipeline(
StandardScaler(), LogisticRegression(solver="liblinear", penalty="l1")
)
results_classification["Logistic Regression L1"] = mean_std_cross_val_scores(
pipe_lgr_l1, X_train, y_train, return_train_score=True, scoring=scoring_metric
)
pd.DataFrame(results_classification)
| dummy | Logistic Regression L2 | Logistic Regression L1 | |
|---|---|---|---|
| fit_time | 0.000 (+/- 0.001) | 0.005 (+/- 0.002) | 0.003 (+/- 0.001) |
| score_time | 0.000 (+/- 0.000) | 0.001 (+/- 0.000) | 0.001 (+/- 0.000) |
| test_score | 0.000 (+/- 0.000) | 0.970 (+/- 0.011) | 0.967 (+/- 0.007) |
| train_score | 0.000 (+/- 0.000) | 0.985 (+/- 0.005) | 0.985 (+/- 0.005) |
The scores are more or less the same.
But L1 regularization is carrying out feature selection; Many coefficients are 0.
Similar scores with less features! More interpretable model!
pipe_lgr_l1.fit(X_train, y_train)
l1_coefs = pipe_lgr_l1.named_steps["logisticregression"].coef_.flatten()
df["l1_coef"] = l1_coefs
df
| l2_coefs | l1_coef | |
|---|---|---|
| mean radius | -0.539854 | 0.000000 |
| mean texture | -0.252141 | 0.000000 |
| mean perimeter | -0.494560 | 0.000000 |
| mean area | -0.610451 | 0.000000 |
| mean smoothness | -0.174066 | 0.000000 |
| mean compactness | 0.577983 | 0.000000 |
| mean concavity | -0.658777 | 0.000000 |
| mean concave points | -0.980244 | -1.337092 |
| mean symmetry | 0.119974 | 0.000000 |
| mean fractal dimension | 0.406479 | 0.271158 |
| radius error | -1.247956 | -2.007079 |
| texture error | -0.103258 | 0.000000 |
| perimeter error | -0.731201 | 0.000000 |
| area error | -0.931584 | 0.000000 |
| smoothness error | -0.230991 | -0.127449 |
| compactness error | 0.614317 | 0.537477 |
| concavity error | -0.032318 | 0.000000 |
| concave points error | -0.206376 | 0.000000 |
| symmetry error | 0.272883 | 0.000000 |
| fractal dimension error | 0.739561 | 0.227465 |
| worst radius | -1.040150 | -1.382266 |
| worst texture | -1.096207 | -1.372887 |
| worst perimeter | -0.885489 | -0.419092 |
| worst area | -1.026863 | -3.613997 |
| worst smoothness | -0.927290 | -0.984068 |
| worst compactness | -0.075403 | 0.000000 |
| worst concavity | -0.766568 | -1.011790 |
| worst concave points | -0.732966 | -0.660896 |
| worst symmetry | -0.669925 | -0.299326 |
| worst fractal dimension | -0.520939 | 0.000000 |
We can also carry out feature selection using L1 regularization and pass selected features to another model.
from lightgbm.sklearn import LGBMClassifier
pipe_lgr_lgbm = make_pipeline(
StandardScaler(),
SelectFromModel(LogisticRegression(solver="liblinear", penalty="l1")),
LGBMClassifier(),
)
results_classification["L1 + LGBM"] = mean_std_cross_val_scores(
pipe_lgr_lgbm,
X_train,
y_train,
return_train_score=True,
scoring=scoring_metric,
)
pd.DataFrame(results_classification)
| dummy | Logistic Regression L2 | Logistic Regression L1 | L1 + LGBM | |
|---|---|---|---|---|
| fit_time | 0.000 (+/- 0.001) | 0.005 (+/- 0.002) | 0.003 (+/- 0.001) | 0.151 (+/- 0.015) |
| score_time | 0.000 (+/- 0.000) | 0.001 (+/- 0.000) | 0.001 (+/- 0.000) | 0.001 (+/- 0.000) |
| test_score | 0.000 (+/- 0.000) | 0.970 (+/- 0.011) | 0.967 (+/- 0.007) | 0.957 (+/- 0.034) |
| train_score | 0.000 (+/- 0.000) | 0.985 (+/- 0.005) | 0.985 (+/- 0.005) | 1.000 (+/- 0.000) |
The score went down a bit this case. But this might help in some other cases.
The resulting model is using L1 selected features only.
How to use regularization with scikit-learn: some examples#
Regression
Least squares with L2-regularization: Ridge
Least squares with L1-regularization: Lasso
Least squares with L1- and L2-regularization: ElasticNet
SVR (\(\epsilon\)-insensitive loss function)
epsilon = 0gives usKernelRidgemodel (least squares with RBF)
Classification
SVC (supports L2-regularization)
LogisticRegression (support L1 and L2 with different solvers)
penalty{‘l1’, ‘l2’, ‘elasticnet’, ‘none’}, default=’l2’ Used to specify the norm used in the penalization. The ‘newton-cg’, ‘sag’ and ‘lbfgs’ solvers support only l2 penalties. ‘elasticnet’ is only supported by the ‘saga’ solver. If ‘none’ (not supported by the liblinear solver), no regularization is applied.
Interpretations of coefficients of regularized linear models should always be taken with a grain of salt.
With different values of the regularization parameter, sometimes even the signs of the coefficients might change!!

Regularization: scaling and collinearity#
Regularization and scaling#
It doesn’t matter for decision trees or naive Bayes.
They only look at one feature at a time.
It doesn’t matter for least squares:
\(w_j*(100 mL)\) gives the same model as \(w_j*(0.1 L)\) with a different \(w_j\)
It matters for \(k\)-nearest neighbours:
Distance will be affected more by large features than small features.
It matters for regularized least squares:
Penalizing \(w_j^2\) means different things if features \(j\) are on different scales
Penalizing \(w_j\) means different things if features \(j\) are on different scales
Collinearity and regularization#
If you have colinear features, the weights would go crazy with regular linear regression.
With L2 regularization: The weight will be equally distributed among all collinear features because the solution is unique.
Example: suppose we have three identical features with a total weight of 1
The weight will be distributed as 1/3, 1/3, 1/3 among the features.
With L1 regularization: The weight will not be equally distributed; the solution is not unique.
Example: suppose we have three identical features with a total weight of 1
The weight could be distributed in many different ways
For example, 1/2, 1/4, 1/4 or 1.0, 0, 0 or 1/2, 1/2, 0 and so on …
Elastic nets#
Combine good properties from both worlds
\(\lambda\) control the strength of regularization
\(\alpha\) controls the amount of sparsity and smoothness
L1 promotes sparsity and the L2 promotes smoothness.
The functional is strictly convex: the solution is unique.
No collinearity problem
A whole group of correlated variables is selected rather than just one variable in the group.
You can use elastic nets using sklearn’s ElasticNet.
Summary: L* regularization#
L0-regularization (AIC, BIC, Mallow’s Cp, Adjusted R2, ANOVA): More on this is DSCi 562
Adds penalty on the number of non-zeros to select features.
L2-regularization (ridge regression):
Adding penalty on the L2-norm of \(w\) to decrease overfitting: $\(f(w) = \lVert Xw - y\rVert_2^2 + \lambda \lVert w\rVert_2^2\)$
L1-regularization (lasso regression):
Adding penalty on the L1-norm decreases overfitting and selects features: $\(f(w) = \lVert Xw - y\rVert_2^2 + \lambda \lVert w\rVert_1\)$
Interpretations of coefficients of linear models should always be taken with a grain of salt
Regularizing a model might change the sign of the coefficients.
Un-regularized linear regression: not affected by scaling
L1 or L2-regularized linear regression: both affected by scaling (and it’s usually a good idea)
