Introduction to covizr
covizr-vignette.RmdThe covizr package provides easy access to Covid-19 data from Our World in Data, as well as functions to generate relevant Covid-19 charts and summaries easily. We aim to make covizr simple and easy to use. Our goal is to enable anyone with basic R programming knowledge to access and visualize Covid-19 data, and make their own informed decisions and conclusions.
We aim to provide simple visualization functions that allow users to answer questions regarding the Covid-19 pandemic as quickly as possible.
This document introduces you to to the basics of using covizr, and shows you how to apply the function.
To use the function, first install the package according to the steps in README, and import the the library with the following code:
library(covizr)Data
To explore the usage of covizr, you do not need extra data. Run the following function as the beginning of the code:
df <- get_data()
df
#> # A tibble: 1,505 × 67
#> iso_code continent location date total_cases new_cases new_cases_smoot…
#> <chr> <chr> <chr> <date> <dbl> <dbl> <dbl>
#> 1 AFG Asia Afghani… 2022-01-29 161290 233 249.
#> 2 AFG Asia Afghani… 2022-01-30 162111 821 352.
#> 3 AFG Asia Afghani… 2022-01-31 162926 815 433.
#> 4 AFG Asia Afghani… 2022-02-01 163555 629 472.
#> 5 AFG Asia Afghani… 2022-02-02 164190 635 500.
#> 6 AFG Asia Afghani… 2022-02-03 164727 537 532.
#> 7 AFG Asia Afghani… 2022-02-04 165358 631 614.
#> 8 ALB Europe Albania 2022-01-29 254126 0 1102
#> 9 ALB Europe Albania 2022-01-30 255741 1615 1096.
#> 10 ALB Europe Albania 2022-01-31 258543 2802 1496.
#> # … with 1,495 more rows, and 60 more variables: total_deaths <dbl>,
#> # new_deaths <dbl>, new_deaths_smoothed <dbl>, total_cases_per_million <dbl>,
#> # new_cases_per_million <dbl>, new_cases_smoothed_per_million <dbl>,
#> # total_deaths_per_million <dbl>, new_deaths_per_million <dbl>,
#> # new_deaths_smoothed_per_million <dbl>, reproduction_rate <dbl>,
#> # icu_patients <dbl>, icu_patients_per_million <dbl>, hosp_patients <dbl>,
#> # hosp_patients_per_million <dbl>, weekly_icu_admissions <dbl>, …By default the function returns the last 7 days of data around the world. There are a few options you can parse into the function for more specific data selection:
- date_from: Start date of the data range with format of ‘YYYY-MM-DD’.
- date_to: End date of the data range with format of ‘YYYY-MM-DD’.
- location: Character vector of target country names.
For example, to retrieve the data between ‘2021-05-01’ to ‘2021-07-01’ of Canada and United Kingdom:
loc <- c('Canada', 'United Kingdom')
df <- get_data(date_from = "2021-05-01", date_to = "2021-07-01", location = loc)
df
#> # A tibble: 124 × 67
#> iso_code continent location date total_cases new_cases new_cases_smoot…
#> <chr> <chr> <chr> <date> <dbl> <dbl> <dbl>
#> 1 CAN North Am… Canada 2021-05-01 1227807 7429 7881.
#> 2 CAN North Am… Canada 2021-05-02 1234733 6926 7891.
#> 3 CAN North Am… Canada 2021-05-03 1243845 9112 7905.
#> 4 CAN North Am… Canada 2021-05-04 1250657 6812 7842.
#> 5 CAN North Am… Canada 2021-05-05 1258014 7357 7790.
#> 6 CAN North Am… Canada 2021-05-06 1266137 8123 7747.
#> 7 CAN North Am… Canada 2021-05-07 1274073 7936 7671.
#> 8 CAN North Am… Canada 2021-05-08 1280731 6658 7561.
#> 9 CAN North Am… Canada 2021-05-09 1287175 6444 7492.
#> 10 CAN North Am… Canada 2021-05-10 1294712 7537 7267.
#> # … with 114 more rows, and 60 more variables: total_deaths <dbl>,
#> # new_deaths <dbl>, new_deaths_smoothed <dbl>, total_cases_per_million <dbl>,
#> # new_cases_per_million <dbl>, new_cases_smoothed_per_million <dbl>,
#> # total_deaths_per_million <dbl>, new_deaths_per_million <dbl>,
#> # new_deaths_smoothed_per_million <dbl>, reproduction_rate <dbl>,
#> # icu_patients <dbl>, icu_patients_per_million <dbl>, hosp_patients <dbl>,
#> # hosp_patients_per_million <dbl>, weekly_icu_admissions <dbl>, …Visualization functions
Summarising a specified variable and value using plot_summary
Use plot_summary when you want to find out what is the top variables (var) from a particular aggregated value (val) or metrics. As with all plotting function in this package, you need to call get_data() first and pass the data frame into the plotting function.
For example, we can see which are the top 10 countries with highest new Covid-19 cases in the first 14 days of 2022 using the following code:
plot_df <- get_data(date_from = "2022-01-01", date_to = "2022-02-01")
plot_summary(plot_df, var = "location", val = "new_cases", date_from = "2022-01-01", date_to = "2022-01-14", top_n = 10)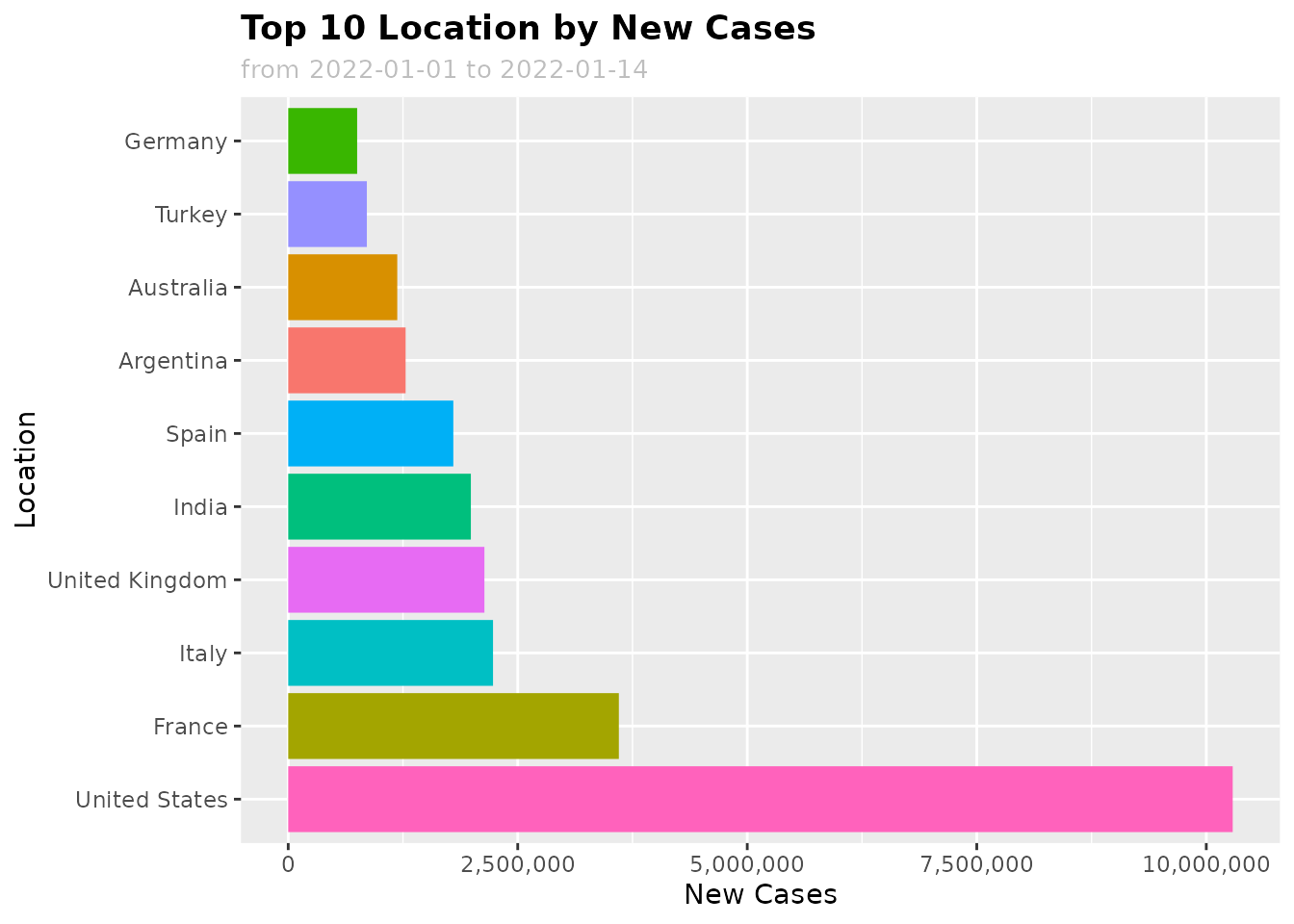
var: Use a categorical variable likelocation,continentval: Use a numeric variable likenew_cases,new_vaccinations,icu_patients
By default, we use sum to aggregate your value of interest. You can use other aggregation function like mean as well, just specify the function name as a string in the argument (e.g. fun = "mean").
Presenting specific countries’ Covid information using plot_spec
We could also get information from specific countries that interest us. By default, this plot_spec function will plot the last 7 days’ new cases trend for Canada. There are a few options you can parse into the function:
- df: Data frame of the selected covid data from get_data(), note that the date range used in get_data function must match or cover the date range here, in order to get complete result
- location: Character vector of target country names.
- val: Quantitative values to be aggregated. Must be numeric variable. Also known as a ‘measure’. Available options including new_deaths, reproduction_rate, icu_patients, hosp_patients, etc.
- date_from: Start date of the data range with format of ‘YYYY-MM-DD’.
- date_to: End date of the data range with format of ‘YYYY-MM-DD’.
- title: The title of the plot.
For example, this plot_spec function below draws the trend of new death cases in Canada and United Kingdom over the first two weeks of January 2022.
df <- get_data(date_from="2022-01-01", date_to="2022-01-15", location = loc)
plot_spec(df, location = c('Canada', 'United Kingdom'), val="new_deaths", date_from="2022-01-01",
date_to="2022-01-15", title="Death cases in Canada and United Kingdom")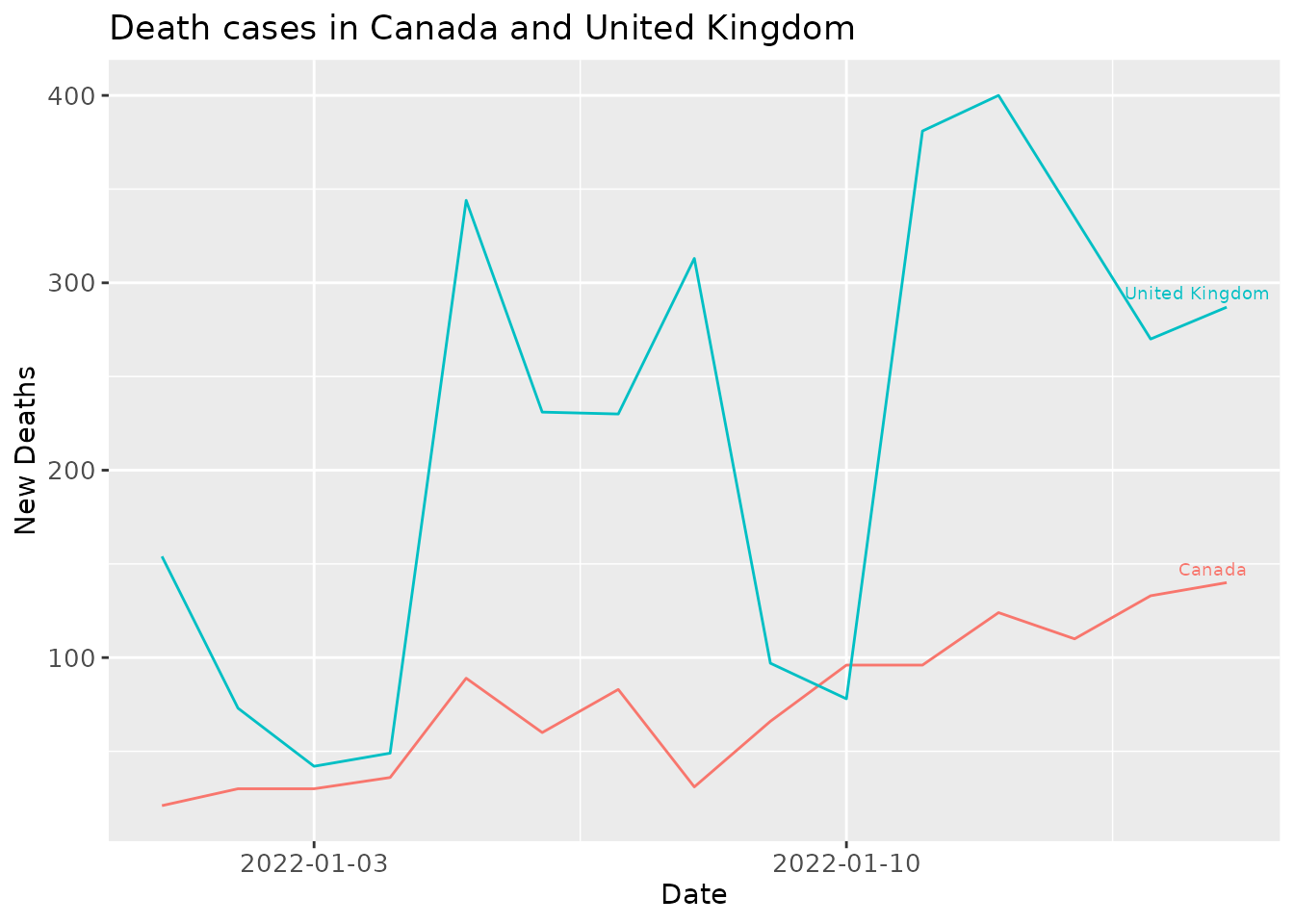
Presenting Covid total new cases verses another metric using plot_metric
After looking at the trend for COVID related cases for specific countries, we can dive deeper into a particular count and visualize the trend of COVID cases with another metric. Some examples of metric which can be used are positive_rate, total_vaccinations, total_deaths, etc. The list of arguments which can be used are provided below:
- df: Data frame of the selected covid data from get_data(), note that the date range used in get_data function must match or cover the date range here, in order to get complete result
- loc_val: Character vector containing a single country.
- metric: Character vector containing a numeric column of the data frame.
- date_from: Start date of the data range with format of ‘YYYY-MM-DD’.
- date_to: End date of the data range with format of ‘YYYY-MM-DD’.
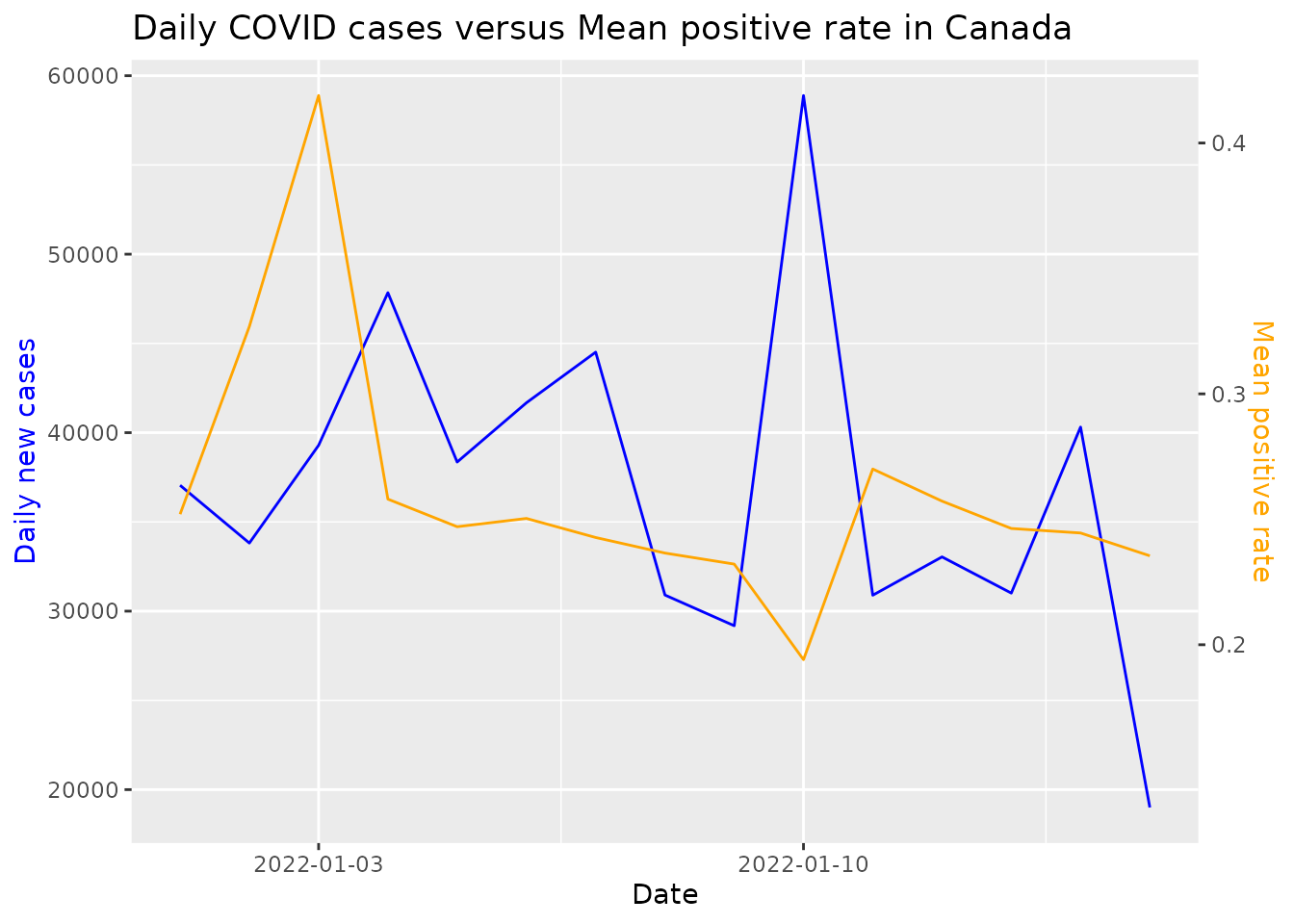
The plot_metric function below charts trend of daily new COVID-19 cases versus the positive rates in Canada for the first two weeks of January 2022.
df <- get_data(date_from="2022-01-01", date_to="2022-01-15", location = c("Canada"))
plot_metric(df, loc_val = c("Canada"), metric = "positive_rate", date_from = "2022-01-01", "2022-01-15")