reasyeda-vignette
reasyeda-vignette.Rmd
library(reasyeda)
## basic example code
library(palmerpenguins) # load the dataset
clean_up(penguins)
#> [1] "**The following potenital outliers are detected:**"
#> $bill_length_mm
#> numeric(0)
#>
#> $bill_depth_mm
#> numeric(0)
#>
#> $flipper_length_mm
#> integer(0)
#>
#> $body_mass_g
#> integer(0)
#>
#> $year
#> integer(0)
#> # A tibble: 333 × 8
#> species island bill_length_mm bill_depth_mm flipper_length_mm body_mass_g
#> <fct> <fct> <dbl> <dbl> <int> <int>
#> 1 Adelie Torgersen 39.1 18.7 181 3750
#> 2 Adelie Torgersen 39.5 17.4 186 3800
#> 3 Adelie Torgersen 40.3 18 195 3250
#> 4 Adelie Torgersen 36.7 19.3 193 3450
#> 5 Adelie Torgersen 39.3 20.6 190 3650
#> 6 Adelie Torgersen 38.9 17.8 181 3625
#> 7 Adelie Torgersen 39.2 19.6 195 4675
#> 8 Adelie Torgersen 41.1 17.6 182 3200
#> 9 Adelie Torgersen 38.6 21.2 191 3800
#> 10 Adelie Torgersen 34.6 21.1 198 4400
#> # … with 323 more rows, and 2 more variables: sex <fct>, year <int>
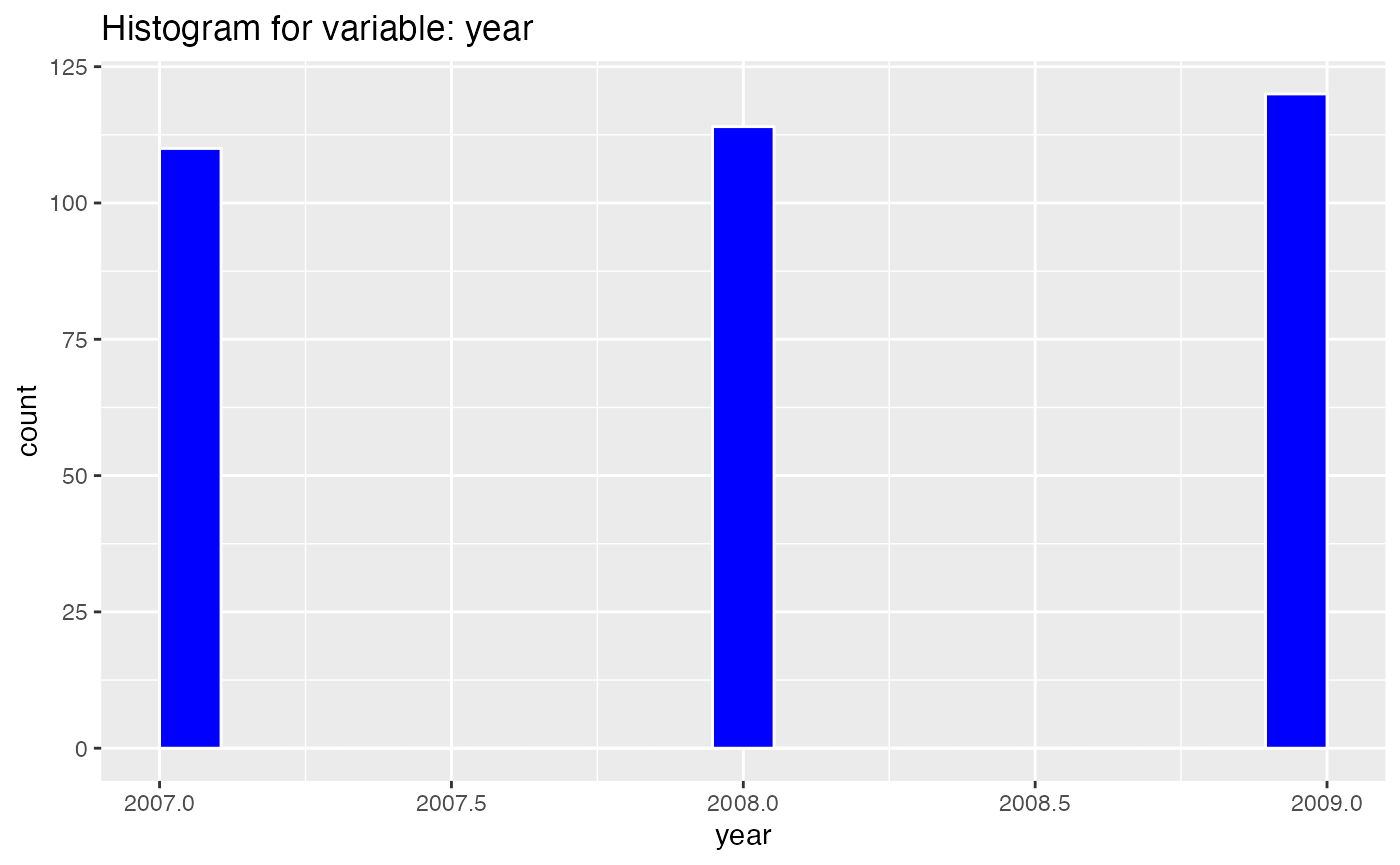
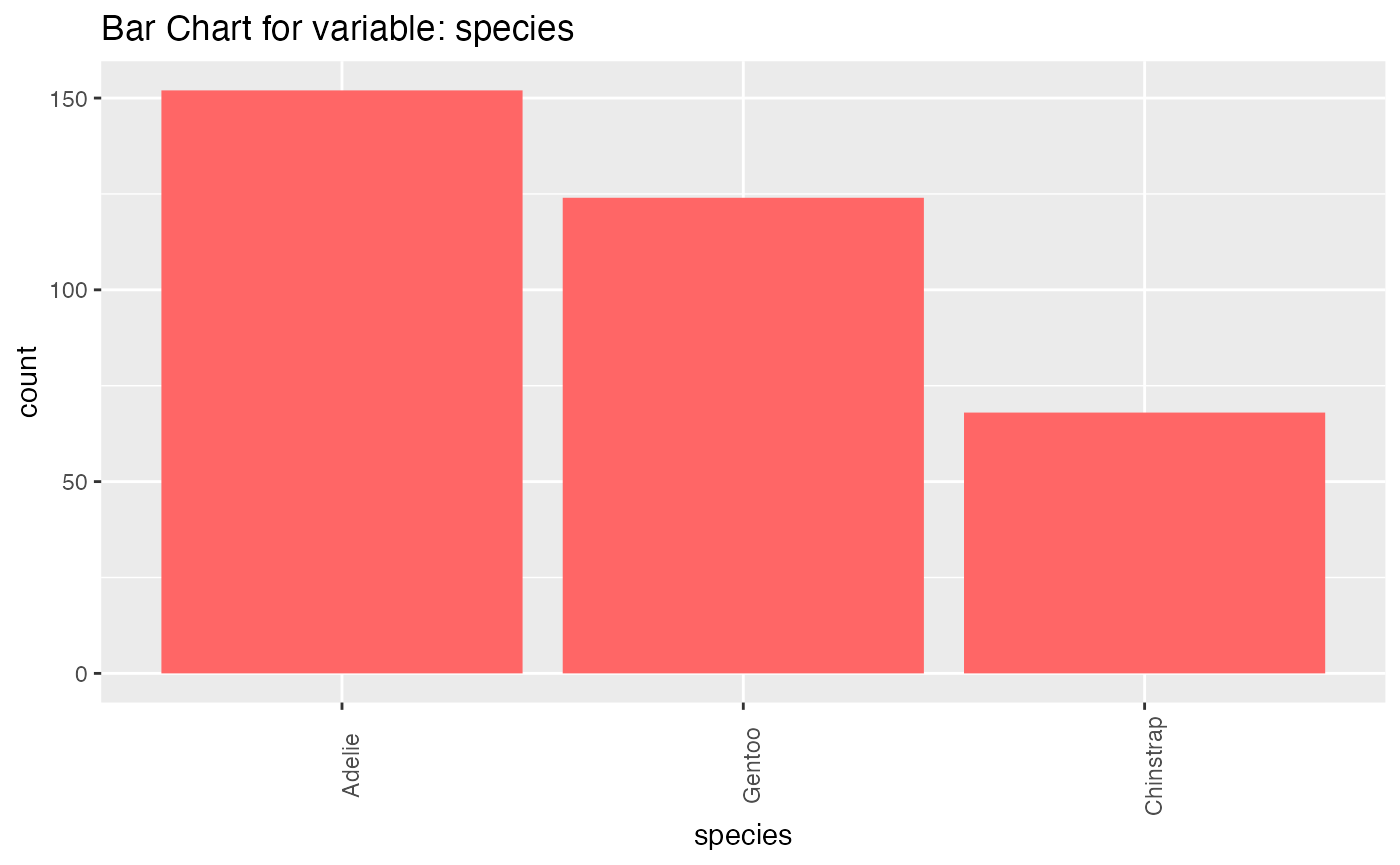
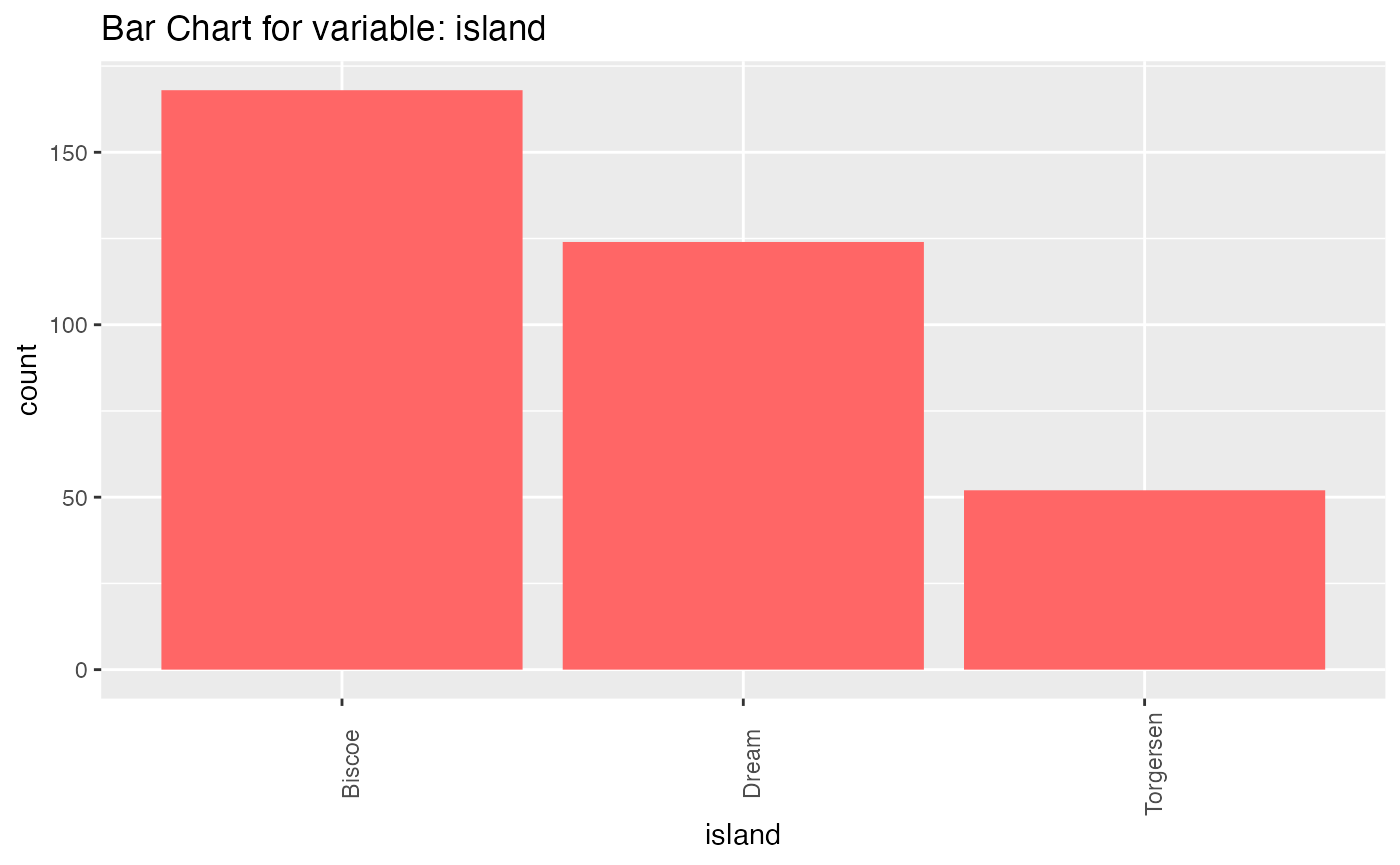
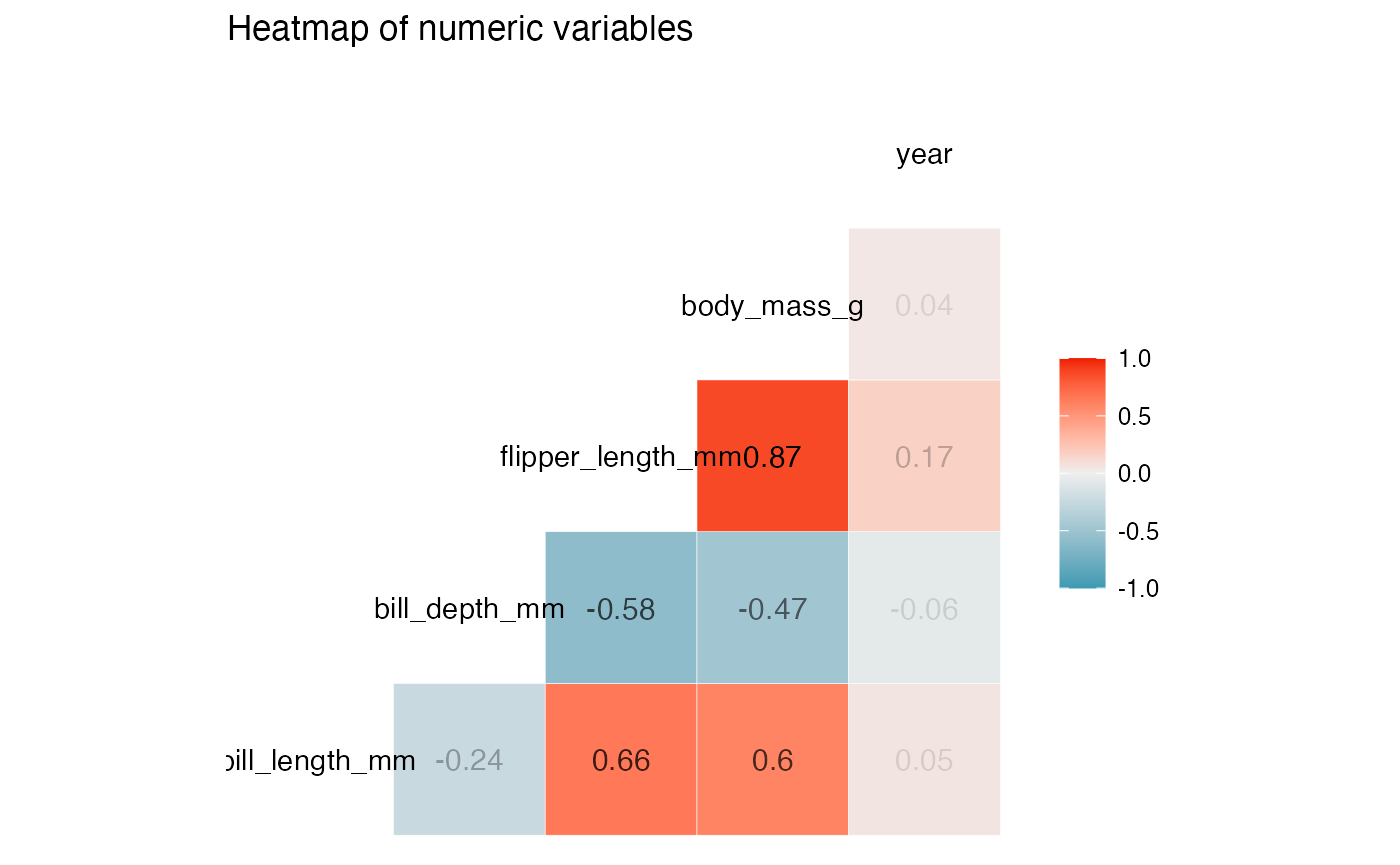
results <- birds_eye_view(penguins)
#> Warning: Removed 2 rows containing non-finite values (stat_bin).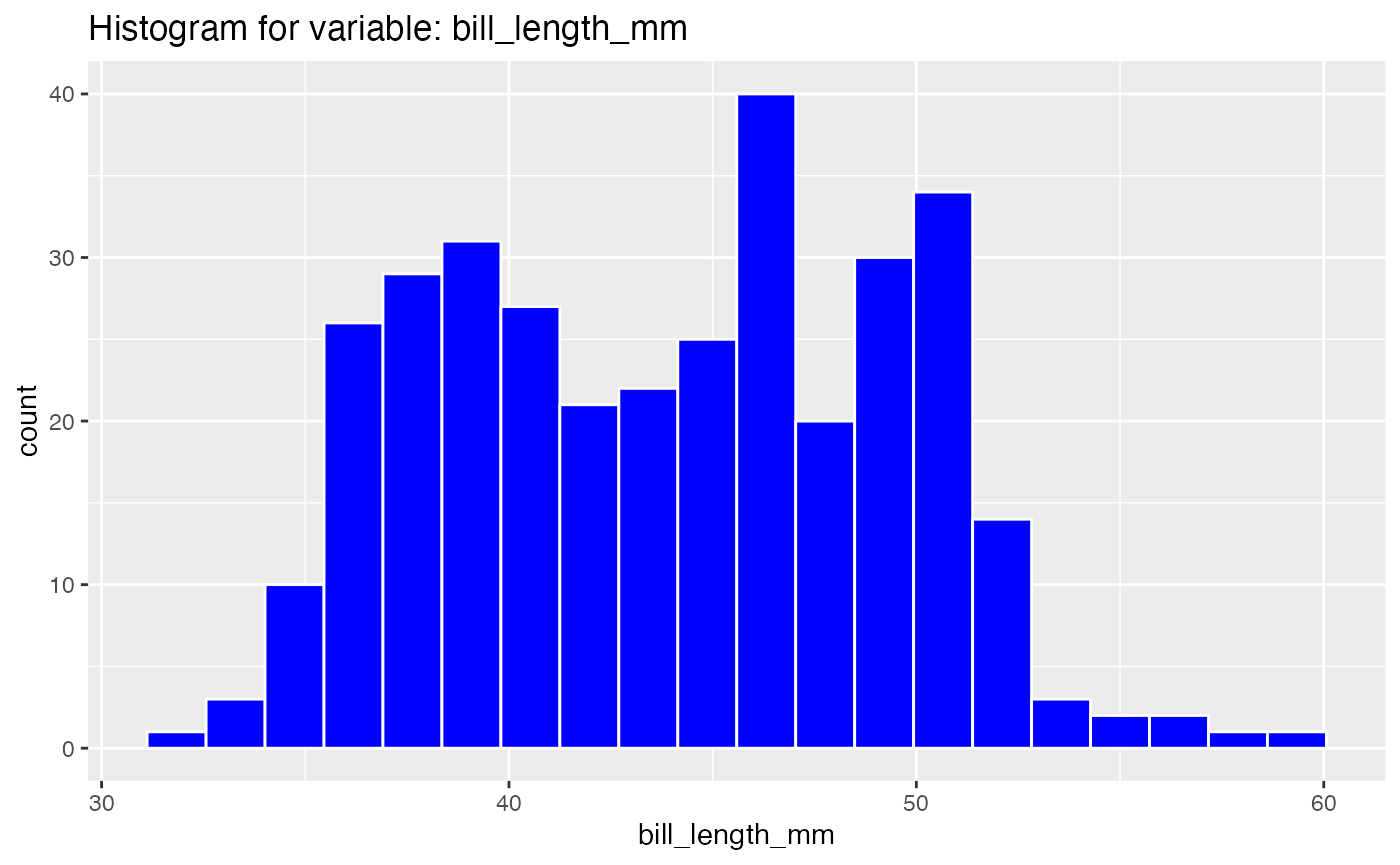
#> Warning: Removed 2 rows containing non-finite values (stat_bin).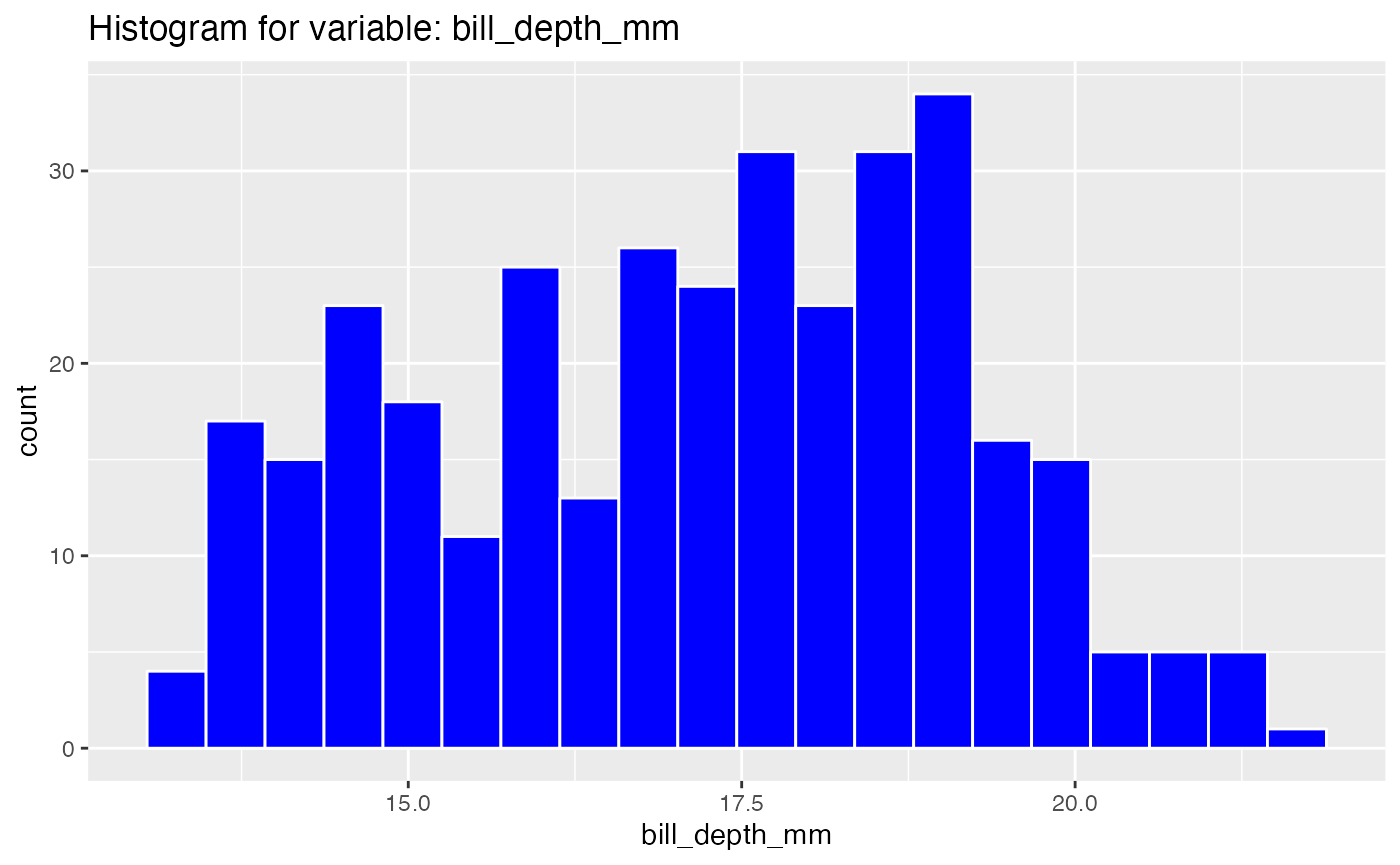
#> Warning: Removed 2 rows containing non-finite values (stat_bin).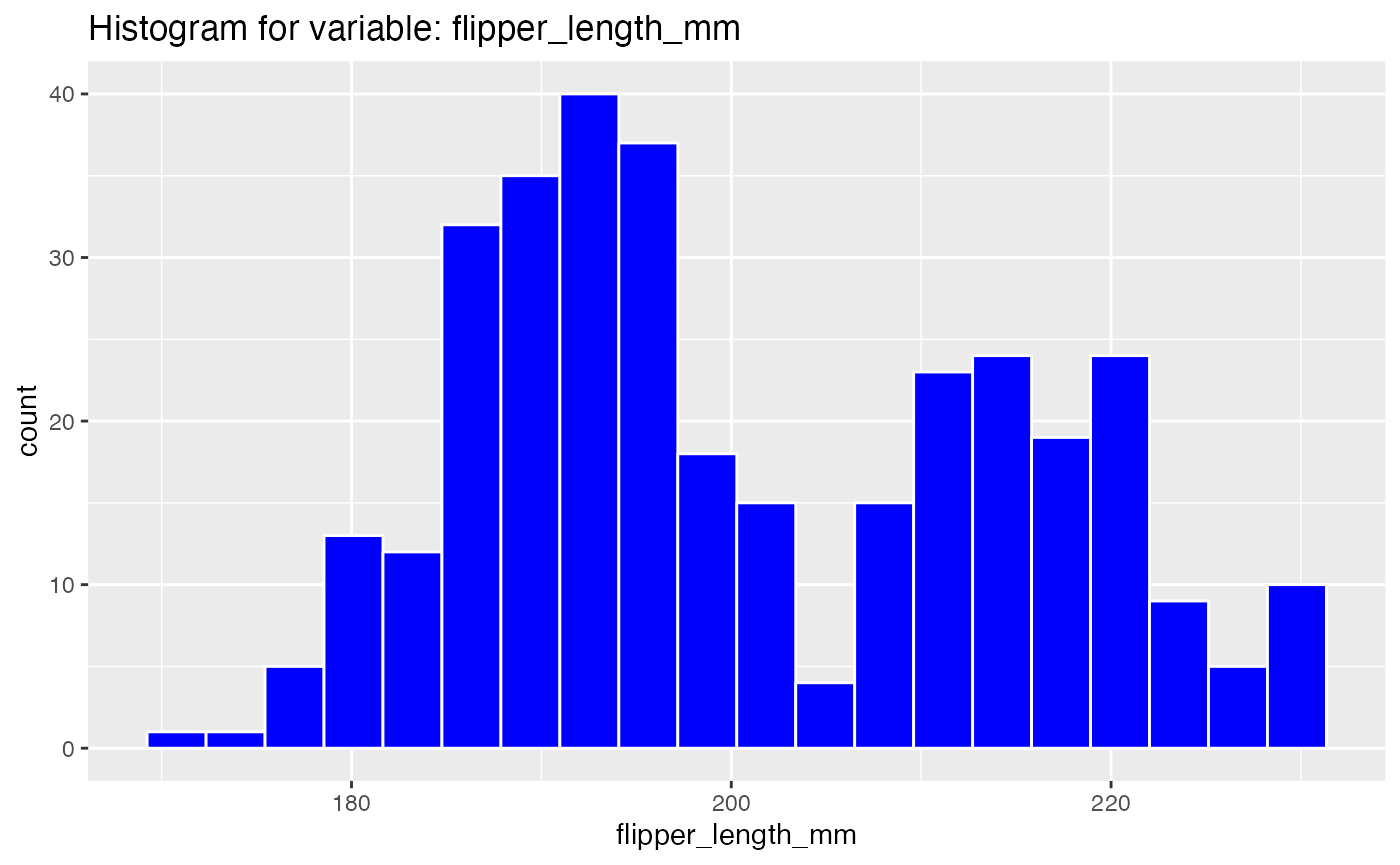
#> Warning: Removed 2 rows containing non-finite values (stat_bin).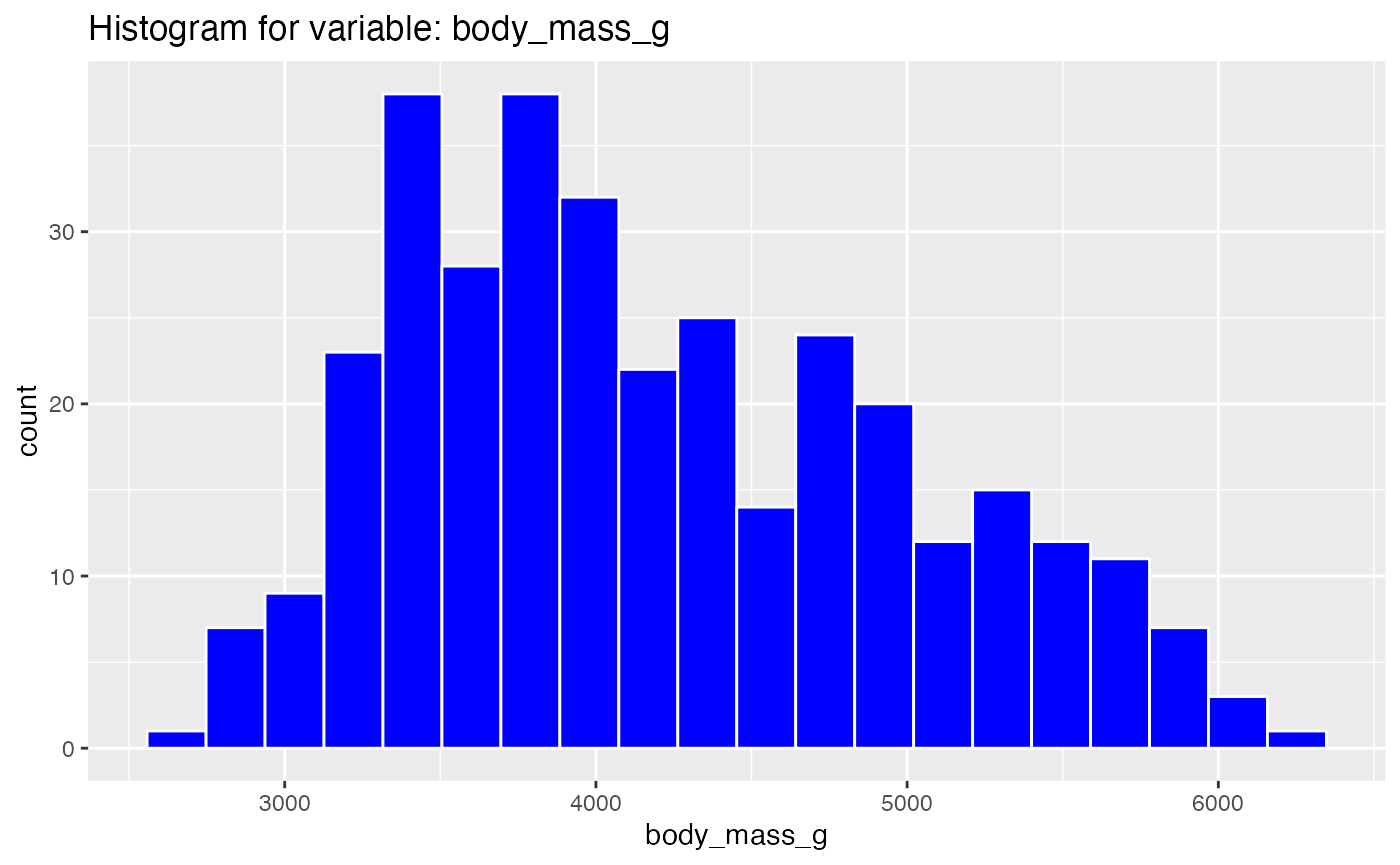
#> Registered S3 method overwritten by 'GGally':
#> method from
#> +.gg ggplot2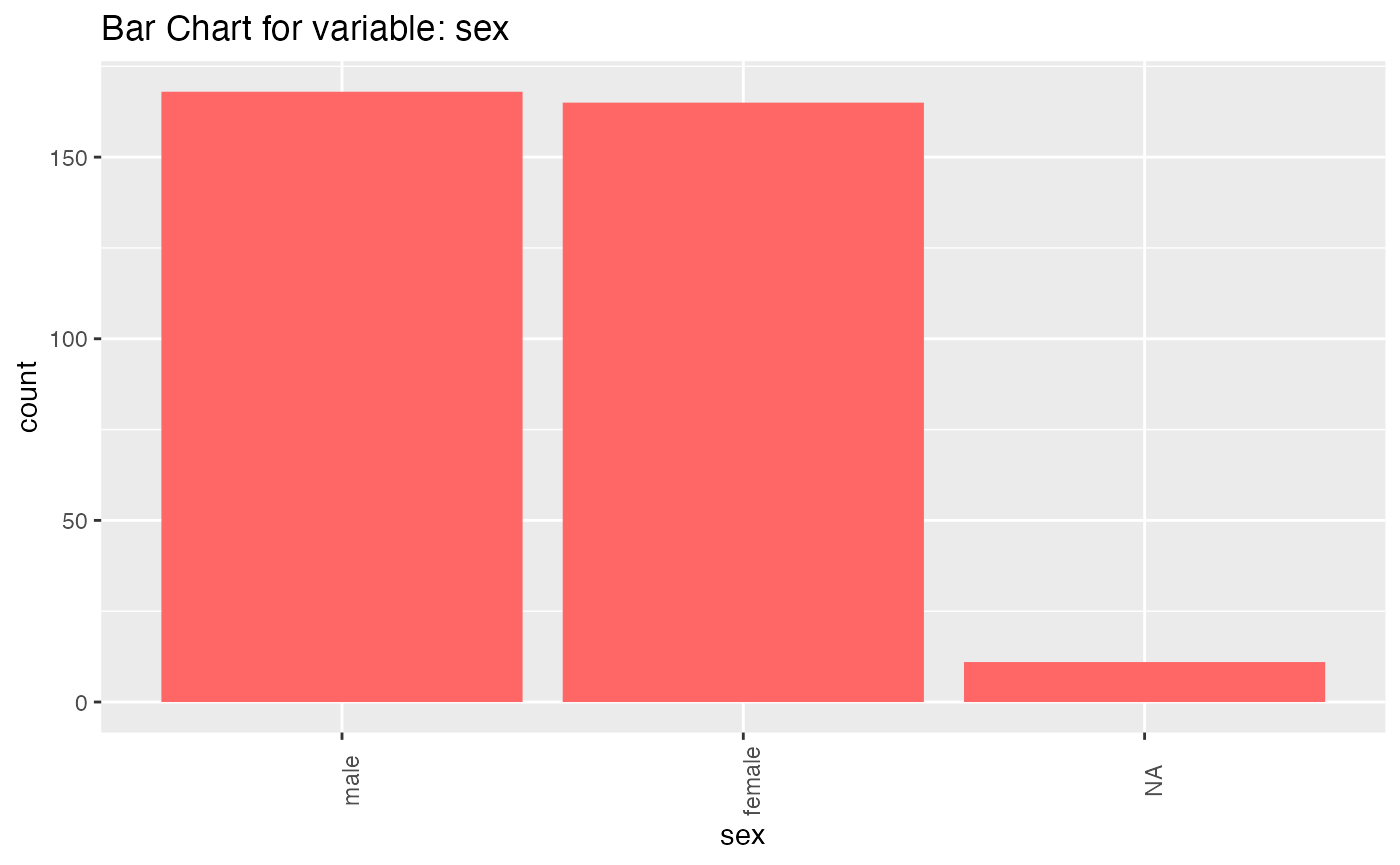
close_up(penguins)
#> Warning: Removed 2 rows containing non-finite values (stat_smooth).
#> Warning: Removed 2 rows containing missing values (geom_point).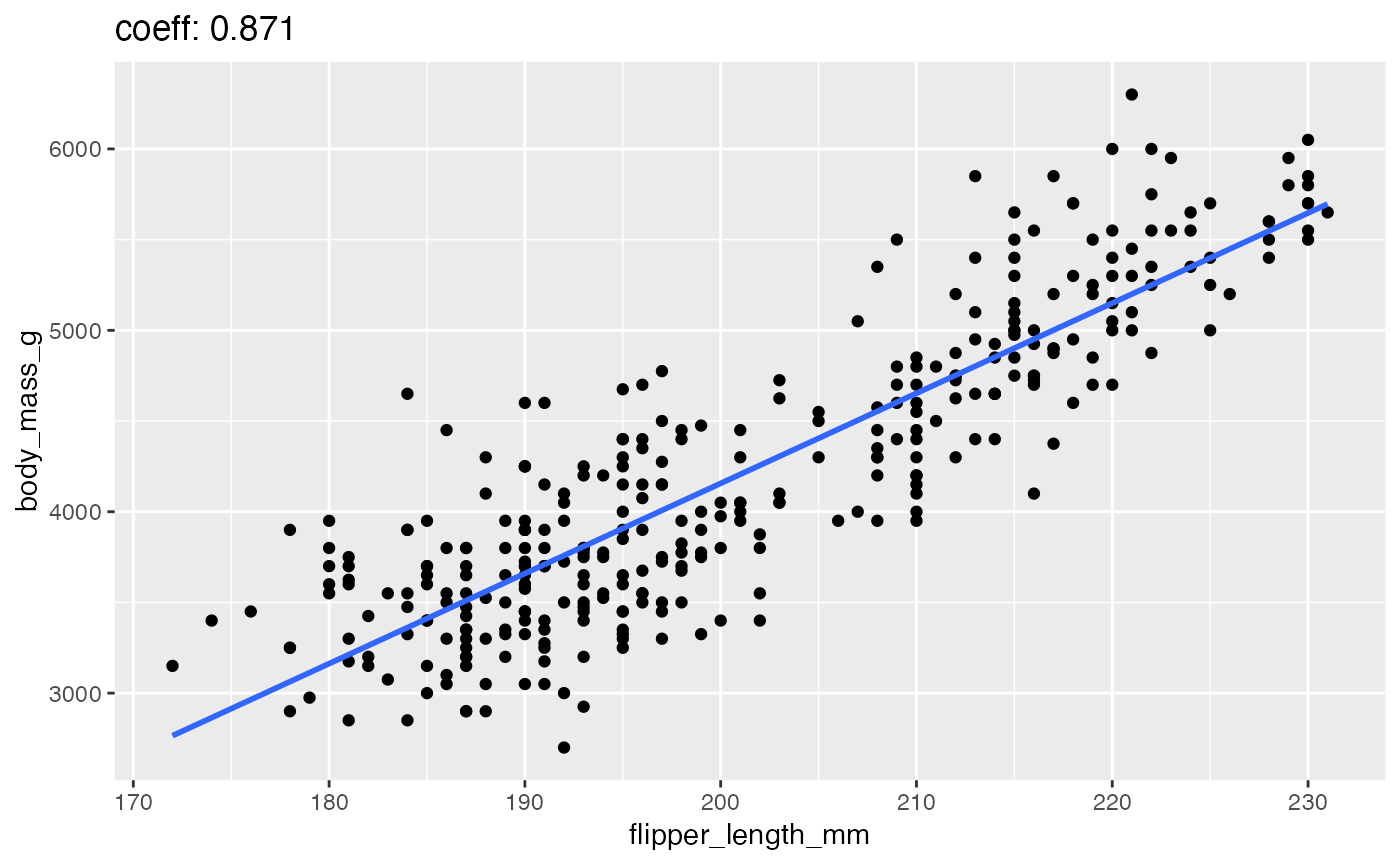
summary_suggestions(penguins)
#> [[1]]
#> bill_length_mm bill_depth_mm flipper_length_mm body_mass_g
#> nbr.val 3.420000e+02 342.0000000 3.420000e+02 3.420000e+02
#> nbr.null 0.000000e+00 0.0000000 0.000000e+00 0.000000e+00
#> nbr.na 2.000000e+00 2.0000000 2.000000e+00 2.000000e+00
#> min 3.210000e+01 13.1000000 1.720000e+02 2.700000e+03
#> max 5.960000e+01 21.5000000 2.310000e+02 6.300000e+03
#> range 2.750000e+01 8.4000000 5.900000e+01 3.600000e+03
#> sum 1.502130e+04 5865.7000000 6.871300e+04 1.437000e+06
#> median 4.445000e+01 17.3000000 1.970000e+02 4.050000e+03
#> mean 4.392193e+01 17.1511696 2.009152e+02 4.201754e+03
#> SE.mean 2.952205e-01 0.1067846 7.603704e-01 4.336473e+01
#> CI.mean.0.95 5.806825e-01 0.2100394 1.495607e+00 8.529605e+01
#> var 2.980705e+01 3.8998080 1.977318e+02 6.431311e+05
#> std.dev 5.459584e+00 1.9747932 1.406171e+01 8.019545e+02
#> coef.var 1.243020e-01 0.1151404 6.998830e-02 1.908618e-01
#> year
#> nbr.val 3.440000e+02
#> nbr.null 0.000000e+00
#> nbr.na 0.000000e+00
#> min 2.007000e+03
#> max 2.009000e+03
#> range 2.000000e+00
#> sum 6.907620e+05
#> median 2.008000e+03
#> mean 2.008029e+03
#> SE.mean 4.412279e-02
#> CI.mean.0.95 8.678531e-02
#> var 6.697064e-01
#> std.dev 8.183559e-01
#> coef.var 4.075419e-04
#>
#> [[2]]
#> dplyr::select_if(df, function(col) is.character(col) | is.factor(col))
#>
#> 3 Variables 344 Observations
#> --------------------------------------------------------------------------------
#> species
#> n missing distinct
#> 344 0 3
#>
#> Value Adelie Chinstrap Gentoo
#> Frequency 152 68 124
#> Proportion 0.442 0.198 0.360
#> --------------------------------------------------------------------------------
#> island
#> n missing distinct
#> 344 0 3
#>
#> Value Biscoe Dream Torgersen
#> Frequency 168 124 52
#> Proportion 0.488 0.360 0.151
#> --------------------------------------------------------------------------------
#> sex
#> n missing distinct
#> 333 11 2
#>
#> Value female male
#> Frequency 165 168
#> Proportion 0.495 0.505
#> --------------------------------------------------------------------------------
#>
#> [[3]]
#> # A tibble: 0 × 0