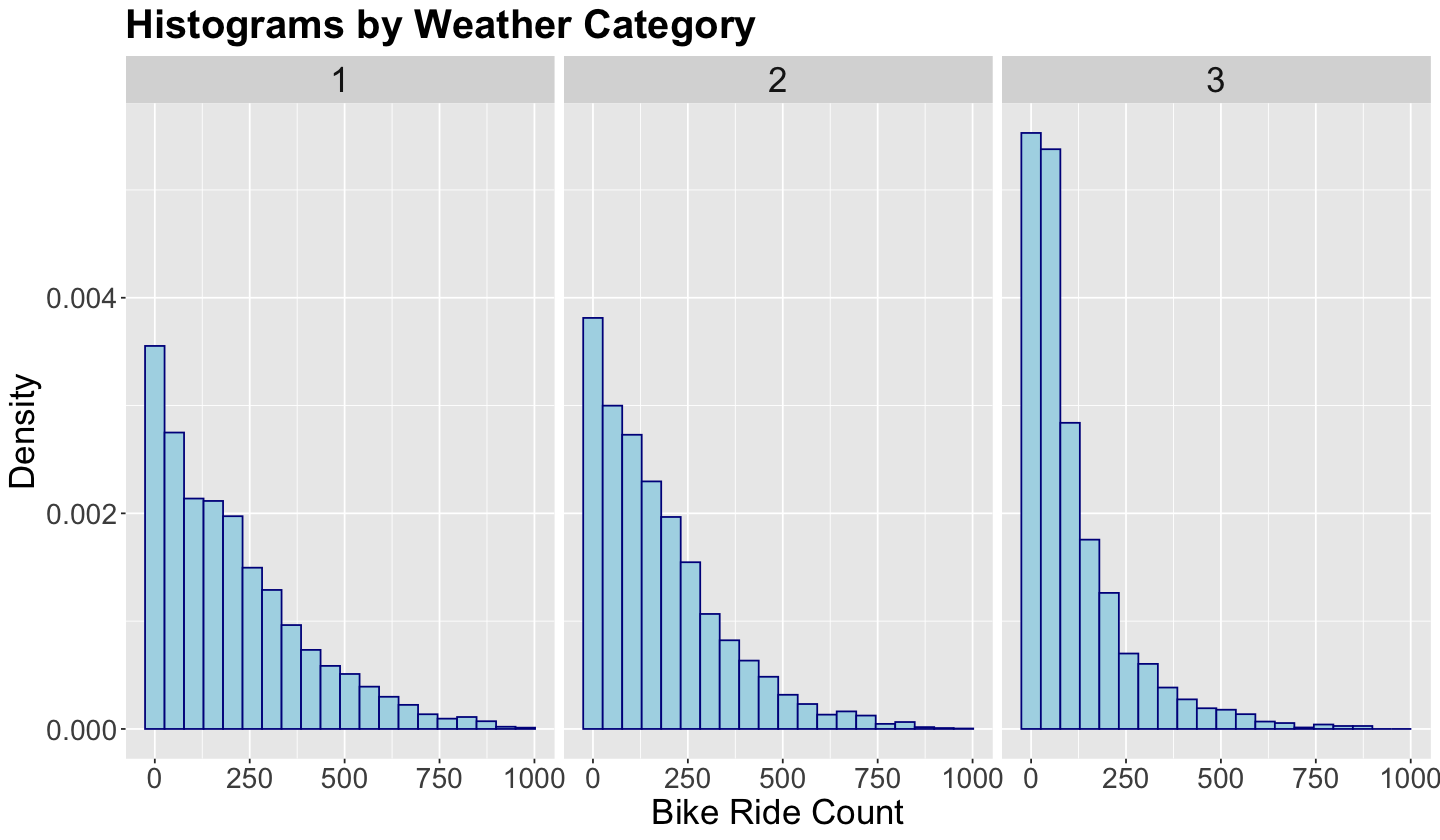
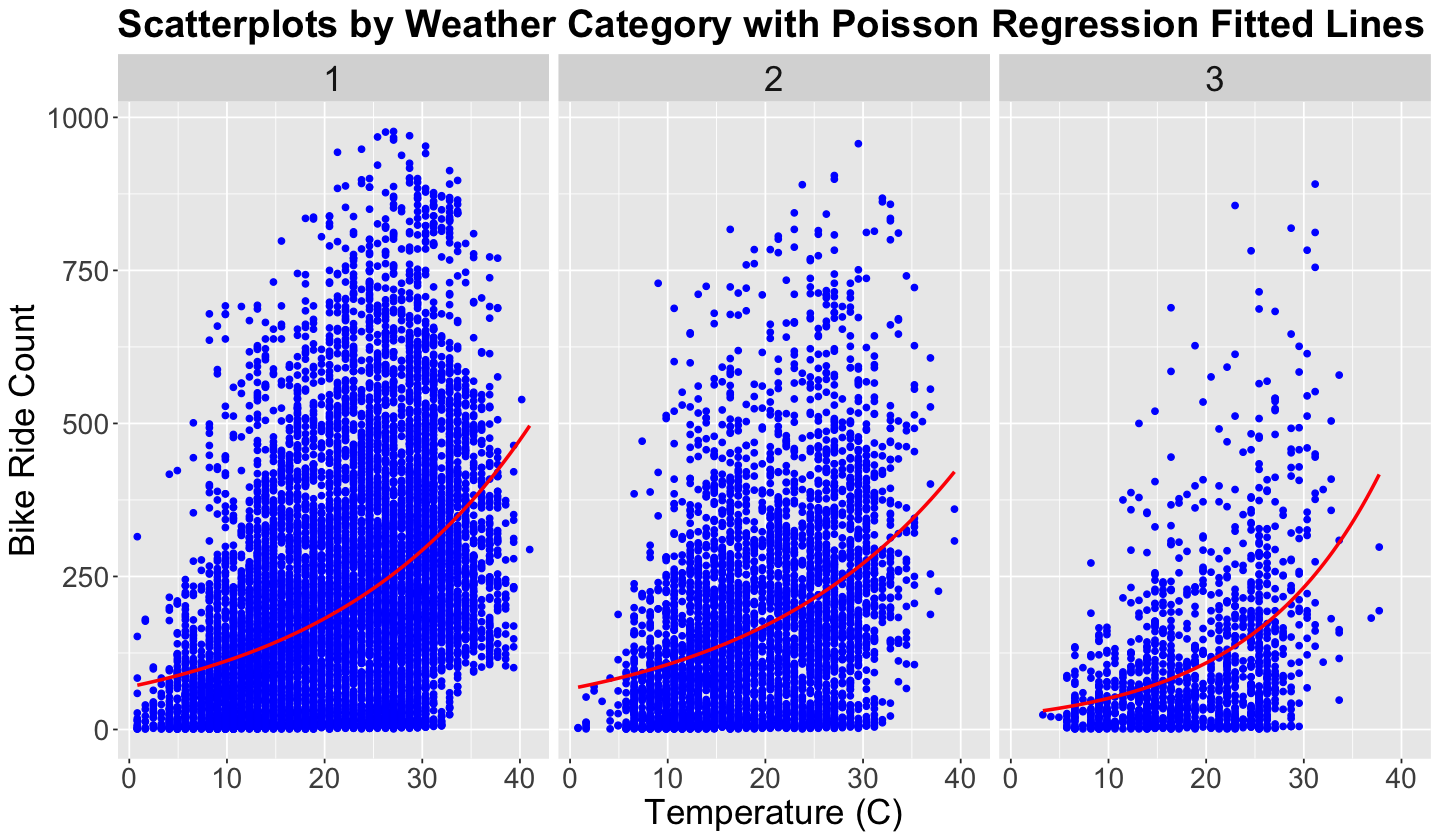
Lecture 4 - Markov Chain Monte Carlo, Stan, and Complex Bayesian Models#
Today’s Learning Objectives#
Illustrate the steps to build a likelihood in a Bayesian design.
Explore a complex dataset involving categorical, count, and continuous variables.
Apply a Bayesian model design with the proper random variables (Normal, Gamma, and Poisson).
Explain the general procedure of Markov Chain Monte Carlo (MCMC).
Practice computational Bayesian inference via
Stan.Draw inferential conclusions from our posterior distribution of parameters.
Loading R Packages#
options(repr.matrix.max.rows = 6, repr.matrix.max.cols = 10)
library(tidyverse)
library(janitor)
library(cowplot)
library(rstan)
library(bayesplot)
library(wesanderson)
Previously…#
We have explored the Bayesian framework on a conceptual level at the beginning of the course. Then, we got familiar with the Bayes’ rule via probability theory. Recall the Bayes’ rule as one of the most powerful components in Bayesian analysis. By applying this rule, we solved a foundational model called the Beta-Binomial in which we combine two random variables via our likelihood and prior distribution. Moreover, we explored the Gamma-Poisson model during lab time.
Nonetheless, real-life problems are not theoretically well behaved as the Beta-Binomial and Gamma Poisson cases. Moreover, complex data will require even more diverse distributions to design our Bayesian model. Hence, the analytical application of the Bayes’ rule will not be possible for a nice exact posterior distribution.
Important
Let us not worry! Stan will come in handy given this issue via MCMC to obtain an approximate posterior distribution.
1. The Big Idea#
Let us recall the big idea of Bayesian modelling. It involves these steps:
Question: Pose a scientific question.
Design: Formulate variables and create a probabilistic model for them. Prior knowledge is included here!
Infer: Get posterior samples from the conditional distribution of any variable you want to infer, given your observed data and prior knowledge.
Check: Make sure the “samples” are actually from your posterior (these are model diagnostics, to be covered later!)
Analyze: Use your samples to compute summaries: mean, maximum a posteriori (MAP), variance, posterior credible intervals, etc.
The scientific question can involve inferential and/or predictive inquiries:
Inferential: Using observed data \(Y\) to infer latent variable \(\theta\).
Predictive: Using observed data \(Y\) to predict future data \(Y'\).
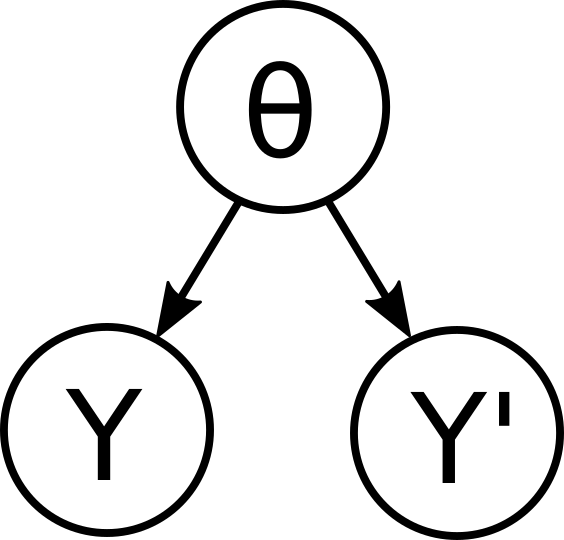
We already discussed their differences in Lecture 3 - Bayesian Statistics in Action: The Beta-Binomial Model our via some examples. Now, let us think generatively!
1.1. To Design the Model, Think Generatively!#
Ultimately, our model is an abstraction of a complex system involving different parameters of interest. Suppose we have \(d\) parameters \(\theta_1, \dots, \theta_d\). Mathematically, these \(d\) parameters can be put in a vector as follows:
Note our Bayesian model is a big joint probability distribution \(P(Y, Y', \Theta)\) over:
Things we will see. Our observations \(Y\).
Things we have to infer: Unobserved parameters \(\Theta\) and predicted observations \(Y'\). This is why the posterior distribution of \(\Theta\) is key!
We already had a first inferential approach (analytically speaking!) via the Beta-Binomial model in Lecture 3 - Bayesian Statistics in Action: The Beta-Binomial Model.
Now, it is much more intuitive to design a couple of steps that generate parameters and synthetic data:
First, generate \(\Theta\) from the prior \(P(\Theta)\).
Then, generate observations \(Y\), \(Y'\) given \(\Theta\) from the likelihood \(P(Y,Y'|\Theta)\).
Attention
We are basically applying the law of conditioning
We will start with the likelihood. This function involves formulating certain variables. It is the core part of the model design! Then, we can establish the prior distributions for our parameters of interest within \(\Theta\) (the likelihood will be of a big help here).
Important
This whole thing is a story of how our data were created.
So far in lab1, we only used our generative Monte Carlo models to simulate synthetic data given some fixed parameters. Nonetheless, we did not incorporate any observed data. In lab2, we will need to include observed data.
1.2. Constructing the Likelihood#
To construct the distribution of the model parameters in \(\Theta\) conditioned on the observed data \(Y\) (i.e., the likelihood), we need to follow these steps:
Step 1: Formulate all data variables - type (integer, real, categorical, etc.) and support (positive, nonnegative, binary, etc.).
Step 2: Figure out which variables we will take as fixed covariates, which will be randomly generated, and how they are related.
Step 3: Pick a distribution with the right support/type/dependencies. The support refers to a proper range of plausible values. Usually, the distribution will have some UNKNOWN PARAMETERS - those will need a prior!
4. Overview of Markov Chain Monte Carlo#
In general, a Bayesian model will have \(d\) parameters of interest for which we will need to obtain a joint posterior distribution:
Recall the Bayes’ rule:
Using \(\Theta\) as our parameter vector of interest, let \(f(\Theta)\) be the multivariate joint prior. Moreover with our observed data \(\mathbf{y} = (y_1, \dots, y_n)^T\), let \(\mathscr{l}(\Theta \mid \mathbf{y})\) be the corresponding multivariate likelihood function along with the multivariate normalizing constant \(f(\mathbf{y})\).
Then, for the multivariate joint posterior \(f(\Theta \mid \mathbf{y})\), the Bayes’ rule becomes:
Why are we highlighting multivariate all the time? It is because we have \(d\) parameters in vector \(\Theta\).
Suppose we want to obtain an analytical form for \(f(\mathbf{y})\), then this will be an integral to solve:
Solving this multivariate integral looks pretty challenging! Nonetheless, recall we do not need to obtain \(f(\mathbf{y})\), but we still need to work around the math in
Important
Cases such as the Beta-Binomial and Gamma-Poisson are well behaved and approachable for parameters \(\pi\) and \(\lambda\), respectively. Therefore, we can obtain a closed solution for the corresponding posterior distribution.
But, what can we do when we have a whole set of parameters in vector \(\Theta\)? It will not be possible to find an exact form for \(f(\Theta \mid \mathbf{y})\), but it is possible to approximate it via an algorithm!
Let us explore the MCMC algorithm by parts: Markov Chain and Monte Carlo.
4.1. Monte Carlo Algorithm#
We will start with the simple Monte Carlo algorithm. Suppose you have a closed analytical form for the posterior distribution \(f(\Theta \mid \mathbf{y})\) (e.g., the Beta posterior in the Beta-Binomial model). You can build your independent Monte Carlo sample \(\{ \Theta_1, \dots, \Theta_N \}\) of size \(N\), from \(f(\Theta \mid \mathbf{y})\) by selecting each element \(\Theta_i\) (\(i = 1, \dots, N\)) in the following way:
Step 1. Draw \(\Theta_i\) from \(f(\Theta \mid \mathbf{y})\).
Step 2. Go there.
This is a random walk!
Caution
Drawing fully independent elements for our sample WILL NOT be possible in MCMC. Moreover, note that \(n \neq N\).
4.2. Markov Chain#
Here comes the MCMC twist to the previous Monte Carlo approach: Markov Chain.
What is a Markov Chain? In terms of our Bayesian modelling for the posterior, it is a sequence \(\{ \Theta_1, \dots, \Theta_N \}\) of size \(N\) whose elements are NOT ENTIRELY INDEPENDENT (hence the word chain!).
What does NOT ENTIRELY INDEPENDENT implicate? It implicates the so-called Markov property:
The next sampled \(\Theta_{i + 1}\) will depend on the current \(\Theta_{i}\) but not on previous \(\Theta_{i - 1}, \Theta_{i - 2}, \dots\)
More formally, because of this property, the conditional probability function of \(\Theta_{i + 1}\) can be depicted as:
Again, there is no free lunch here! Each element in this chain can be drawn from another distribution and not specifically from our target posterior \(f(\Theta \mid \mathbf{y})\):
Thus, what is the point of this Markov Chain approach? The answer lies in the MCMC algorithm that approximates \(f(\Theta \mid \mathbf{y})\) via our \(N\) elements in the chain.
Important
There is more than one MCMC algorithm to do so, and the foundational one is the Metropolis-Hastings algorithm.
4.3. The Metropolis-Hastings Algorithm#
This is the algorithm that will allow us to obtain an approximation of the posterior distribution \(f(\Theta \mid \mathbf{y})\) via our Markov Chain \(\{ \Theta_1, \dots, \Theta_N \}\). Hence, with \(\Theta_i\) in our current iteration \((i = 1, 2, \dots, N - 1)\), the next iteration \(\Theta_{i + 1}\) will be selected through a two-step process.
Recall the Bayes’ rule:
Let the current \(\Theta_i = \Theta\). Then, the algorithm’s two steps are:
Step 1. Randomly draw a new location \(\Theta'\) from a proposed model with PDF \(q(\Theta' \mid \Theta)\). This proposed model is what we call a jumping distribution and it could be a Uniform or Normal which are symmetric.
Important
Another choice of a jumping distribution could be asymmetric, such as the Log-normal. We can choose this jumping distribution class when there are constraints on our parameters of interest (e.g., they are nonnegative), or we want to sample large values from the chain (recall the Log-normal is a heavy-tailed distribution).
Step 2. Now, let us decide to go or not to go to the proposed \(\Theta'\):
Important
Step 2 is divided in two parts:
a. Compute the acceptance probability for \(\Theta'\):
b. Obtain the next location \(\Theta_{i + 1}\) as a Bernoulli trial (flipping an unfair coin!) taking on the two possible values:
4.4. The Metropolis Algorithm#
The previous Metropolis-Hastings algorithm can be simplified when the PDF of the jumping distribution is Uniform such that:
This jumping distribution allocates the same probability of switching from \(\Theta\) to \(\Theta'\) compared to switching from \(\Theta'\) to \(\Theta\). Note that \(w\) is the window parameter. Having said that, the definition of \(\alpha\) will change to:
4.5. Other MCMC Algorithms#
Metropolis-Hastings and its simplified Metropolis version are only a couple of MCMC tools. These two approaches target single parameters (e.g., a probability of success \(\pi\) or a mean \(\lambda\)). However, when we want to deal with \(d\) parameters, such as \(\Theta = (\theta_1, \dots, \theta_d)^T\), there are more complex tools like Gibbs sampling (used in JAGS, Just Another Gibbs Sampler) or the No-U-Turn sampler (NUTS) which is a Hamiltonian Monte Carlo (HMC) algorithm (implemented in Stan).
4.6. Important Facts to Consider when sampling() via rstan#
The MCMC algorithm has a learning curve; thus, the Markov Chain initially starts sampling unreasonable numbers to approximate the posterior \(f(\Theta \mid \mathbf{y})\). Therefore, we have a burn-in stage at the beginning of sampling() defined by the parameter warmup. This parameter will discard the first warmup iterations for the final simulation output.
Recall that consecutive elements in the chain are not independent. Hence, given the Markov property, we could skip draws between our definitive sampled values throughout the simulation to achieve a certain degree of sampling independence. This is defined by thin.
Important
We will obtain the following number of approximate posterior samples via sampling():
5. Using Stan to Obtain our Posterior Distribution#
We already designed our Bayesian model. Now it is time to code it in Stan. Recall that with a dataset containing \(n\) observations (i.e., rows in bikeshare_data which is our training set size), for the \(i\)th observation \((i = 1, \dots, n)\) with temperature \(t_i\):
5.1. Coding the Model#
Recall Stan has a block structure. Therefore, we will use the following blocks:
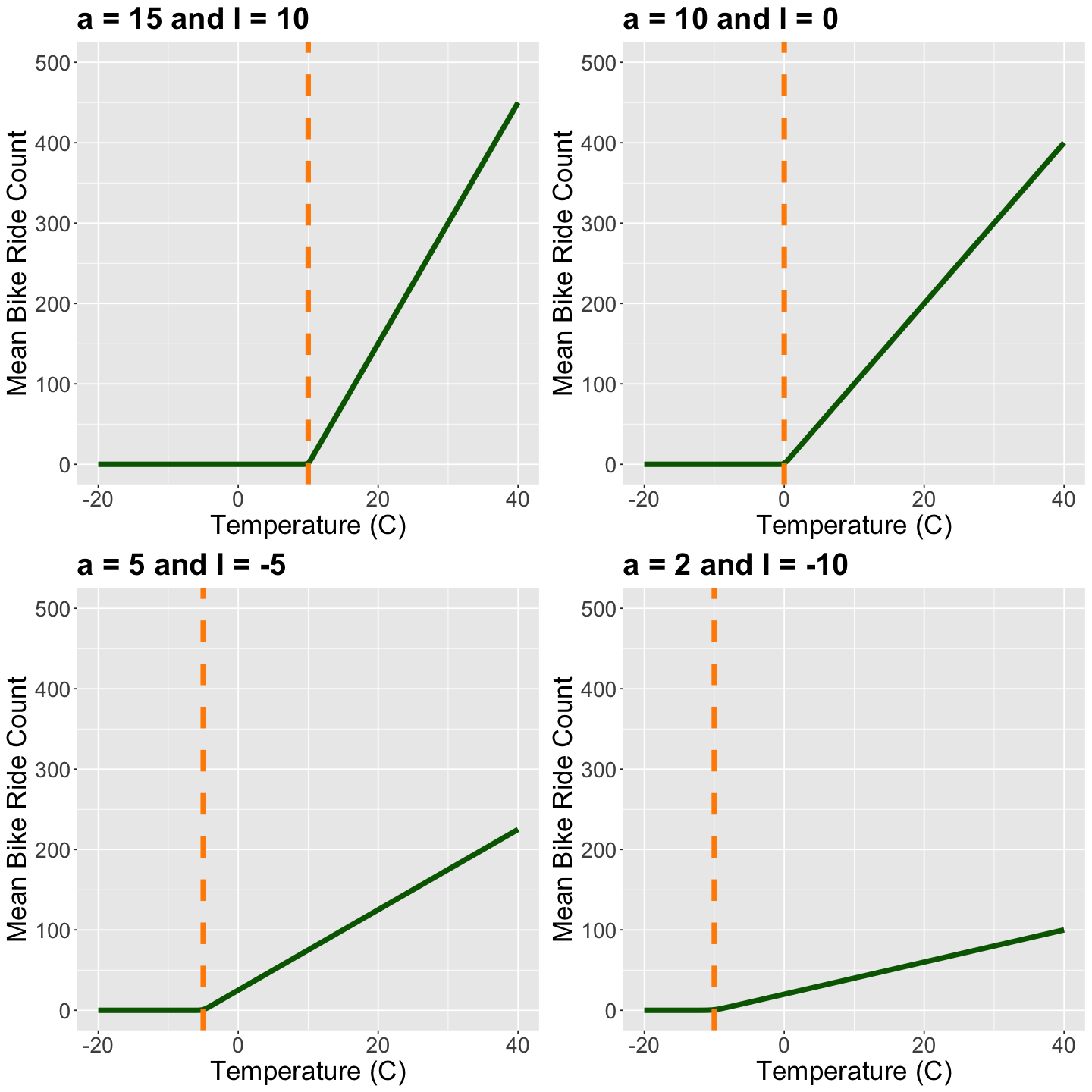
data: The inputs needed to run the Bayesian model.parameters: The parameters to be sampled for our posterior analysis (\(a_w\) and \(l_w\) for \(w \in \{1, 2, 3\}\)). Their approximate posterior distributions will allow us to establish the extent to which temperature and mean bike ride count are related across different categories of weather (this is Bayesian inference!).model: Our Bayesian model.
Caution
Stan needs to know every single detail of our data and generative model (object types, sizes, loops, etc.).
Unlike the R markdown, we do not have a Stan cell in Jupyter notebook/lab. Hence, we need to code our bikeshare_model as a string.
bikeshare_model <- "data {
int<lower=1> number_weather_cat; // integer to indicate how many weather categories we have
int<lower=1> n; // how many rows we have in bikeshare_data
vector[n] temp; // column bikeshare_data$temp of real type
int<lower=1> weather[n]; // column bikeshare_data$weather as integer
int<lower=0> ride_count[n]; // column bikeshare_data$ride_count as integer
}
parameters {
vector<lower=0>[number_weather_cat] a; // a vector of size number_weather_cat for a_w
vector[number_weather_cat] l; // a real-type vector of size number_weather_cat for l_w
}
model {
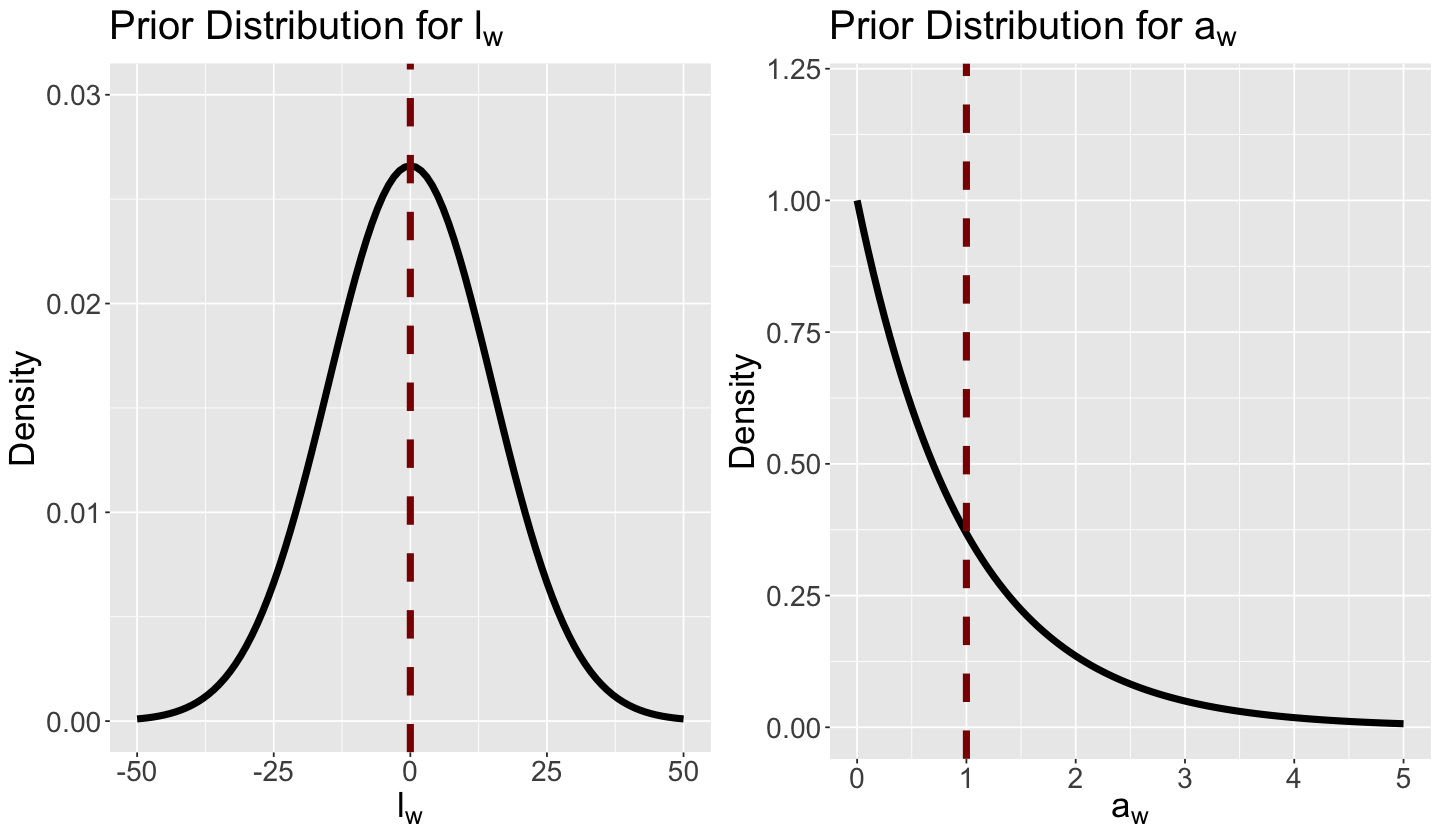
for (j in 1:number_weather_cat){ // we have three PAIRS of priors, i.e., one PAIR per weather category
a[j] ~ gamma(1, 1);
l[j] ~ normal(0, 15);
}
for (i in 1:n){ // building our likelihood
int w_index = weather[i]; // auxiliary variable for the ith weather category
ride_count[i] ~ poisson(log(1 + exp(a[w_index] * (temp[i] - l[w_index]))));
}
}"
5.2. MCMC Simulation#
Next, we create a dictionary with all the data variables we need to pass into Stan as a list().
bikeshare_dictionary <- list(
number_weather_cat = 3,
n = nrow(bikeshare_data),
temp = bikeshare_data$temp,
weather = as.integer(bikeshare_data$weather),
ride_count = as.integer(bikeshare_data$ride_count)
)
We have not compiled bikeshare_model. Hence, we cannot use sampling() directly but stan() as in the cell below. Note the MCMC arguments are provided as in sampling():
chains: How many times we want to run the Bayesian simulation.iter: The number of approximate posterior sampled sets ofparameterswe want (including burn-in).warmup: The burn-in period.thin: Number of samples to skip between each draw.seed: Reproducibility seed.
posterior_bikeshare <- stan(
model_code = bikeshare_model,
data = bikeshare_dictionary,
chains = 1,
iter = 20000,
warmup = 2000,
thin = 40,
seed = 553,
verbose = TRUE,
algorithm = "NUTS"
)
Show code cell output
TRANSLATING MODEL '' FROM Stan CODE TO C++ CODE NOW.
OS: aarch64, darwin20; rstan: 2.32.6; Rcpp: 1.0.12; inline: 0.3.19
>> setting environment variables:
PKG_LIBS = '/Library/Frameworks/R.framework/Versions/4.4-arm64/Resources/library/rstan/lib//libStanServices.a' -L'/Library/Frameworks/R.framework/Versions/4.4-arm64/Resources/library/StanHeaders/lib/' -lStanHeaders -L'/Library/Frameworks/R.framework/Versions/4.4-arm64/Resources/library/RcppParallel/lib/' -ltbb -L/Library/Frameworks/R.framework/Versions/4.4-arm64/Resources/library/RcppParallel/lib -ltbb -ltbbmalloc
PKG_CPPFLAGS = -I"/Library/Frameworks/R.framework/Versions/4.4-arm64/Resources/library/Rcpp/include/" -I"/Library/Frameworks/R.framework/Versions/4.4-arm64/Resources/library/RcppEigen/include/" -I"/Library/Frameworks/R.framework/Versions/4.4-arm64/Resources/library/RcppEigen/include/unsupported" -I"/Library/Frameworks/R.framework/Versions/4.4-arm64/Resources/library/BH/include" -I"/Library/Frameworks/R.framework/Versions/4.4-arm64/Resources/library/StanHeaders/include/src/" -I"/Library/Frameworks/R.framework/Versions/4.4-arm64/Resources/library/StanHeaders/include/" -I"/Library/Frameworks/R.framework/Versions/4.4-arm64/Resources/library/RcppParallel/include/" -I"/Library/Frameworks/R.framework/Versions/4.4-arm64/Resources/library/rstan/include" -DEIGEN_NO_DEBUG -DBOOST_DISABLE_ASSERTS -DBOOST_PENDING_INTEGER_LOG2_HPP -DSTAN_THREADS -DUSE_STANC3 -DSTRICT_R_HEADERS -DBOOST_PHOENIX_NO_VARIADIC_EXPRESSION -D_HAS_AUTO_PTR_ETC=0 -include '/Library/Frameworks/R.framework/Versions/4.4-arm64/Resources/library/StanHeaders/include/stan/math/prim/fun/Eigen.hpp' -D_REENTRANT -DRCPP_PARALLEL_USE_TBB=1
>> Program source :
1 :
2 : // includes from the plugin
3 : // [[Rcpp::plugins(cpp14)]]
4 :
5 :
6 : // user includes
7 : #include <Rcpp.h>
8 : using namespace Rcpp;
9 : #ifndef MODELS_HPP
10 : #define MODELS_HPP
11 : #define STAN__SERVICES__COMMAND_HPP
12 : #include <rstan/rstaninc.hpp>
13 : #ifndef USE_STANC3
14 : #define USE_STANC3
15 : #endif
16 : // Code generated by stanc v2.32.2
17 : #include <stan/model/model_header.hpp>
18 : namespace model18558745e4a24__namespace {
19 : using stan::model::model_base_crtp;
20 : using namespace stan::math;
21 : stan::math::profile_map profiles__;
22 : static constexpr std::array<const char*, 21> locations_array__ =
23 : {" (found before start of program)",
24 : " (in 'anon_model', line 9, column 0 to column 38)",
25 : " (in 'anon_model', line 10, column 0 to column 29)",
26 : " (in 'anon_model', line 14, column 2 to column 21)",
27 : " (in 'anon_model', line 15, column 2 to column 23)",
28 : " (in 'anon_model', line 13, column 31 to line 16, column 1)",
29 : " (in 'anon_model', line 13, column 0 to line 16, column 1)",
30 : " (in 'anon_model', line 18, column 2 to column 27)",
31 : " (in 'anon_model', line 19, column 2 to column 77)",
32 : " (in 'anon_model', line 17, column 14 to line 20, column 1)",
33 : " (in 'anon_model', line 17, column 0 to line 20, column 1)",
34 : " (in 'anon_model', line 2, column 0 to column 32)",
35 : " (in 'anon_model', line 3, column 0 to column 15)",
36 : " (in 'anon_model', line 4, column 7 to column 8)",
37 : " (in 'anon_model', line 4, column 0 to column 15)",
38 : " (in 'anon_model', line 5, column 21 to column 22)",
39 : " (in 'anon_model', line 5, column 0 to column 24)",
40 : " (in 'anon_model', line 6, column 24 to column 25)",
41 : " (in 'anon_model', line 6, column 0 to column 27)",
42 : " (in 'anon_model', line 9, column 16 to column 34)",
43 : " (in 'anon_model', line 10, column 7 to column 25)"};
44 : class model18558745e4a24_ final : public model_base_crtp<model18558745e4a24_> {
45 : private:
46 : int number_weather_cat;
47 : int n;
48 : Eigen::Matrix<double,-1,1> temp_data__;
49 : std::vector<int> weather;
50 : std::vector<int> ride_count;
51 : Eigen::Map<Eigen::Matrix<double,-1,1>> temp{nullptr, 0};
52 : public:
53 : ~model18558745e4a24_() {}
54 : model18558745e4a24_(stan::io::var_context& context__, unsigned int
55 : random_seed__ = 0, std::ostream* pstream__ = nullptr)
56 : : model_base_crtp(0) {
57 : int current_statement__ = 0;
58 : using local_scalar_t__ = double;
59 : boost::ecuyer1988 base_rng__ =
60 : stan::services::util::create_rng(random_seed__, 0);
61 : // suppress unused var warning
62 : (void) base_rng__;
63 : static constexpr const char* function__ =
64 : "model18558745e4a24__namespace::model18558745e4a24_";
65 : // suppress unused var warning
66 : (void) function__;
67 : local_scalar_t__ DUMMY_VAR__(std::numeric_limits<double>::quiet_NaN());
68 : // suppress unused var warning
69 : (void) DUMMY_VAR__;
70 : try {
71 : int pos__ = std::numeric_limits<int>::min();
72 : pos__ = 1;
73 : current_statement__ = 11;
74 : context__.validate_dims("data initialization", "number_weather_cat",
75 : "int", std::vector<size_t>{});
76 : number_weather_cat = std::numeric_limits<int>::min();
77 : current_statement__ = 11;
78 : number_weather_cat = context__.vals_i("number_weather_cat")[(1 - 1)];
79 : current_statement__ = 11;
80 : stan::math::check_greater_or_equal(function__, "number_weather_cat",
81 : number_weather_cat, 1);
82 : current_statement__ = 12;
83 : context__.validate_dims("data initialization", "n", "int",
84 : std::vector<size_t>{});
85 : n = std::numeric_limits<int>::min();
86 : current_statement__ = 12;
87 : n = context__.vals_i("n")[(1 - 1)];
88 : current_statement__ = 12;
89 : stan::math::check_greater_or_equal(function__, "n", n, 1);
90 : current_statement__ = 13;
91 : stan::math::validate_non_negative_index("temp", "n", n);
92 : current_statement__ = 14;
93 : context__.validate_dims("data initialization", "temp", "double",
94 : std::vector<size_t>{static_cast<size_t>(n)});
95 : temp_data__ = Eigen::Matrix<double,-1,1>::Constant(n,
96 : std::numeric_limits<double>::quiet_NaN());
97 : new (&temp) Eigen::Map<Eigen::Matrix<double,-1,1>>(temp_data__.data(),
98 : n);
99 : {
100 : std::vector<local_scalar_t__> temp_flat__;
101 : current_statement__ = 14;
102 : temp_flat__ = context__.vals_r("temp");
103 : current_statement__ = 14;
104 : pos__ = 1;
105 : current_statement__ = 14;
106 : for (int sym1__ = 1; sym1__ <= n; ++sym1__) {
107 : current_statement__ = 14;
108 : stan::model::assign(temp, temp_flat__[(pos__ - 1)],
109 : "assigning variable temp", stan::model::index_uni(sym1__));
110 : current_statement__ = 14;
111 : pos__ = (pos__ + 1);
112 : }
113 : }
114 : current_statement__ = 15;
115 : stan::math::validate_non_negative_index("weather", "n", n);
116 : current_statement__ = 16;
117 : context__.validate_dims("data initialization", "weather", "int",
118 : std::vector<size_t>{static_cast<size_t>(n)});
119 : weather = std::vector<int>(n, std::numeric_limits<int>::min());
120 : current_statement__ = 16;
121 : weather = context__.vals_i("weather");
122 : current_statement__ = 16;
123 : stan::math::check_greater_or_equal(function__, "weather", weather, 1);
124 : current_statement__ = 17;
125 : stan::math::validate_non_negative_index("ride_count", "n", n);
126 : current_statement__ = 18;
127 : context__.validate_dims("data initialization", "ride_count", "int",
128 : std::vector<size_t>{static_cast<size_t>(n)});
129 : ride_count = std::vector<int>(n, std::numeric_limits<int>::min());
130 : current_statement__ = 18;
131 : ride_count = context__.vals_i("ride_count");
132 : current_statement__ = 18;
133 : stan::math::check_greater_or_equal(function__, "ride_count",
134 : ride_count, 0);
135 : current_statement__ = 19;
136 : stan::math::validate_non_negative_index("a", "number_weather_cat",
137 : number_weather_cat);
138 : current_statement__ = 20;
139 : stan::math::validate_non_negative_index("l", "number_weather_cat",
140 : number_weather_cat);
141 : } catch (const std::exception& e) {
142 : stan::lang::rethrow_located(e, locations_array__[current_statement__]);
143 : }
144 : num_params_r__ = number_weather_cat + number_weather_cat;
145 : }
146 : inline std::string model_name() const final {
147 : return "model18558745e4a24_";
148 : }
149 : inline std::vector<std::string> model_compile_info() const noexcept {
150 : return std::vector<std::string>{"stanc_version = stanc3 v2.32.2",
151 : "stancflags = --"};
152 : }
153 : template <bool propto__, bool jacobian__, typename VecR, typename VecI,
154 : stan::require_vector_like_t<VecR>* = nullptr,
155 : stan::require_vector_like_vt<std::is_integral, VecI>* = nullptr>
156 : inline stan::scalar_type_t<VecR>
157 : log_prob_impl(VecR& params_r__, VecI& params_i__, std::ostream*
158 : pstream__ = nullptr) const {
159 : using T__ = stan::scalar_type_t<VecR>;
160 : using local_scalar_t__ = T__;
161 : T__ lp__(0.0);
162 : stan::math::accumulator<T__> lp_accum__;
163 : stan::io::deserializer<local_scalar_t__> in__(params_r__, params_i__);
164 : int current_statement__ = 0;
165 : local_scalar_t__ DUMMY_VAR__(std::numeric_limits<double>::quiet_NaN());
166 : // suppress unused var warning
167 : (void) DUMMY_VAR__;
168 : static constexpr const char* function__ =
169 : "model18558745e4a24__namespace::log_prob";
170 : // suppress unused var warning
171 : (void) function__;
172 : try {
173 : Eigen::Matrix<local_scalar_t__,-1,1> a =
174 : Eigen::Matrix<local_scalar_t__,-1,1>::Constant(number_weather_cat,
175 : DUMMY_VAR__);
176 : current_statement__ = 1;
177 : a = in__.template read_constrain_lb<
178 : Eigen::Matrix<local_scalar_t__,-1,1>, jacobian__>(0, lp__,
179 : number_weather_cat);
180 : Eigen::Matrix<local_scalar_t__,-1,1> l =
181 : Eigen::Matrix<local_scalar_t__,-1,1>::Constant(number_weather_cat,
182 : DUMMY_VAR__);
183 : current_statement__ = 2;
184 : l = in__.template read<
185 : Eigen::Matrix<local_scalar_t__,-1,1>>(number_weather_cat);
186 : {
187 : current_statement__ = 6;
188 : for (int j = 1; j <= number_weather_cat; ++j) {
189 : current_statement__ = 3;
190 : lp_accum__.add(stan::math::gamma_lpdf<propto__>(
191 : stan::model::rvalue(a, "a",
192 : stan::model::index_uni(j)), 1, 1));
193 : current_statement__ = 4;
194 : lp_accum__.add(stan::math::normal_lpdf<propto__>(
195 : stan::model::rvalue(l, "l",
196 : stan::model::index_uni(j)), 0, 15));
197 : }
198 : current_statement__ = 10;
199 : for (int i = 1; i <= n; ++i) {
200 : int w_index = std::numeric_limits<int>::min();
201 : current_statement__ = 7;
202 : w_index = stan::model::rvalue(weather, "weather",
203 : stan::model::index_uni(i));
204 : current_statement__ = 8;
205 : lp_accum__.add(stan::math::poisson_lpmf<propto__>(
206 : stan::model::rvalue(ride_count, "ride_count",
207 : stan::model::index_uni(i)),
208 : stan::math::log((1 +
209 : stan::math::exp(
210 : (stan::model::rvalue(a, "a",
211 : stan::model::index_uni(w_index)) *
212 : (stan::model::rvalue(temp, "temp",
213 : stan::model::index_uni(i)) -
214 : stan::model::rvalue(l, "l",
215 : stan::model::index_uni(w_index)))))))));
216 : }
217 : }
218 : } catch (const std::exception& e) {
219 : stan::lang::rethrow_located(e, locations_array__[current_statement__]);
220 : }
221 : lp_accum__.add(lp__);
222 : return lp_accum__.sum();
223 : }
224 : template <typename RNG, typename VecR, typename VecI, typename VecVar,
225 : stan::require_vector_like_vt<std::is_floating_point,
226 : VecR>* = nullptr, stan::require_vector_like_vt<std::is_integral,
227 : VecI>* = nullptr, stan::require_vector_vt<std::is_floating_point,
228 : VecVar>* = nullptr>
229 : inline void
230 : write_array_impl(RNG& base_rng__, VecR& params_r__, VecI& params_i__,
231 : VecVar& vars__, const bool
232 : emit_transformed_parameters__ = true, const bool
233 : emit_generated_quantities__ = true, std::ostream*
234 : pstream__ = nullptr) const {
235 : using local_scalar_t__ = double;
236 : stan::io::deserializer<local_scalar_t__> in__(params_r__, params_i__);
237 : stan::io::serializer<local_scalar_t__> out__(vars__);
238 : static constexpr bool propto__ = true;
239 : // suppress unused var warning
240 : (void) propto__;
241 : double lp__ = 0.0;
242 : // suppress unused var warning
243 : (void) lp__;
244 : int current_statement__ = 0;
245 : stan::math::accumulator<double> lp_accum__;
246 : local_scalar_t__ DUMMY_VAR__(std::numeric_limits<double>::quiet_NaN());
247 : // suppress unused var warning
248 : (void) DUMMY_VAR__;
249 : constexpr bool jacobian__ = false;
250 : static constexpr const char* function__ =
251 : "model18558745e4a24__namespace::write_array";
252 : // suppress unused var warning
253 : (void) function__;
254 : try {
255 : Eigen::Matrix<double,-1,1> a =
256 : Eigen::Matrix<double,-1,1>::Constant(number_weather_cat,
257 : std::numeric_limits<double>::quiet_NaN());
258 : current_statement__ = 1;
259 : a = in__.template read_constrain_lb<
260 : Eigen::Matrix<local_scalar_t__,-1,1>, jacobian__>(0, lp__,
261 : number_weather_cat);
262 : Eigen::Matrix<double,-1,1> l =
263 : Eigen::Matrix<double,-1,1>::Constant(number_weather_cat,
264 : std::numeric_limits<double>::quiet_NaN());
265 : current_statement__ = 2;
266 : l = in__.template read<
267 : Eigen::Matrix<local_scalar_t__,-1,1>>(number_weather_cat);
268 : out__.write(a);
269 : out__.write(l);
270 : if (stan::math::logical_negation(
271 : (stan::math::primitive_value(emit_transformed_parameters__) ||
272 : stan::math::primitive_value(emit_generated_quantities__)))) {
273 : return ;
274 : }
275 : if (stan::math::logical_negation(emit_generated_quantities__)) {
276 : return ;
277 : }
278 : } catch (const std::exception& e) {
279 : stan::lang::rethrow_located(e, locations_array__[current_statement__]);
280 : }
281 : }
282 : template <typename VecVar, typename VecI,
283 : stan::require_vector_t<VecVar>* = nullptr,
284 : stan::require_vector_like_vt<std::is_integral, VecI>* = nullptr>
285 : inline void
286 : unconstrain_array_impl(const VecVar& params_r__, const VecI& params_i__,
287 : VecVar& vars__, std::ostream* pstream__ = nullptr) const {
288 : using local_scalar_t__ = double;
289 : stan::io::deserializer<local_scalar_t__> in__(params_r__, params_i__);
290 : stan::io::serializer<local_scalar_t__> out__(vars__);
291 : int current_statement__ = 0;
292 : local_scalar_t__ DUMMY_VAR__(std::numeric_limits<double>::quiet_NaN());
293 : // suppress unused var warning
294 : (void) DUMMY_VAR__;
295 : try {
296 : int pos__ = std::numeric_limits<int>::min();
297 : pos__ = 1;
298 : Eigen::Matrix<local_scalar_t__,-1,1> a =
299 : Eigen::Matrix<local_scalar_t__,-1,1>::Constant(number_weather_cat,
300 : DUMMY_VAR__);
301 : current_statement__ = 1;
302 : stan::model::assign(a,
303 : in__.read<Eigen::Matrix<local_scalar_t__,-1,1>>(number_weather_cat),
304 : "assigning variable a");
305 : out__.write_free_lb(0, a);
306 : Eigen::Matrix<local_scalar_t__,-1,1> l =
307 : Eigen::Matrix<local_scalar_t__,-1,1>::Constant(number_weather_cat,
308 : DUMMY_VAR__);
309 : current_statement__ = 2;
310 : stan::model::assign(l,
311 : in__.read<Eigen::Matrix<local_scalar_t__,-1,1>>(number_weather_cat),
312 : "assigning variable l");
313 : out__.write(l);
314 : } catch (const std::exception& e) {
315 : stan::lang::rethrow_located(e, locations_array__[current_statement__]);
316 : }
317 : }
318 : template <typename VecVar, stan::require_vector_t<VecVar>* = nullptr>
319 : inline void
320 : transform_inits_impl(const stan::io::var_context& context__, VecVar&
321 : vars__, std::ostream* pstream__ = nullptr) const {
322 : using local_scalar_t__ = double;
323 : stan::io::serializer<local_scalar_t__> out__(vars__);
324 : int current_statement__ = 0;
325 : local_scalar_t__ DUMMY_VAR__(std::numeric_limits<double>::quiet_NaN());
326 : // suppress unused var warning
327 : (void) DUMMY_VAR__;
328 : try {
329 : current_statement__ = 1;
330 : context__.validate_dims("parameter initialization", "a", "double",
331 : std::vector<size_t>{static_cast<size_t>(number_weather_cat)});
332 : current_statement__ = 2;
333 : context__.validate_dims("parameter initialization", "l", "double",
334 : std::vector<size_t>{static_cast<size_t>(number_weather_cat)});
335 : int pos__ = std::numeric_limits<int>::min();
336 : pos__ = 1;
337 : Eigen::Matrix<local_scalar_t__,-1,1> a =
338 : Eigen::Matrix<local_scalar_t__,-1,1>::Constant(number_weather_cat,
339 : DUMMY_VAR__);
340 : {
341 : std::vector<local_scalar_t__> a_flat__;
342 : current_statement__ = 1;
343 : a_flat__ = context__.vals_r("a");
344 : current_statement__ = 1;
345 : pos__ = 1;
346 : current_statement__ = 1;
347 : for (int sym1__ = 1; sym1__ <= number_weather_cat; ++sym1__) {
348 : current_statement__ = 1;
349 : stan::model::assign(a, a_flat__[(pos__ - 1)],
350 : "assigning variable a", stan::model::index_uni(sym1__));
351 : current_statement__ = 1;
352 : pos__ = (pos__ + 1);
353 : }
354 : }
355 : out__.write_free_lb(0, a);
356 : Eigen::Matrix<local_scalar_t__,-1,1> l =
357 : Eigen::Matrix<local_scalar_t__,-1,1>::Constant(number_weather_cat,
358 : DUMMY_VAR__);
359 : {
360 : std::vector<local_scalar_t__> l_flat__;
361 : current_statement__ = 2;
362 : l_flat__ = context__.vals_r("l");
363 : current_statement__ = 2;
364 : pos__ = 1;
365 : current_statement__ = 2;
366 : for (int sym1__ = 1; sym1__ <= number_weather_cat; ++sym1__) {
367 : current_statement__ = 2;
368 : stan::model::assign(l, l_flat__[(pos__ - 1)],
369 : "assigning variable l", stan::model::index_uni(sym1__));
370 : current_statement__ = 2;
371 : pos__ = (pos__ + 1);
372 : }
373 : }
374 : out__.write(l);
375 : } catch (const std::exception& e) {
376 : stan::lang::rethrow_located(e, locations_array__[current_statement__]);
377 : }
378 : }
379 : inline void
380 : get_param_names(std::vector<std::string>& names__, const bool
381 : emit_transformed_parameters__ = true, const bool
382 : emit_generated_quantities__ = true) const {
383 : names__ = std::vector<std::string>{"a", "l"};
384 : if (emit_transformed_parameters__) {}
385 : if (emit_generated_quantities__) {}
386 : }
387 : inline void
388 : get_dims(std::vector<std::vector<size_t>>& dimss__, const bool
389 : emit_transformed_parameters__ = true, const bool
390 : emit_generated_quantities__ = true) const {
391 : dimss__ = std::vector<std::vector<size_t>>{std::vector<size_t>{static_cast<
392 : size_t>(
393 : number_weather_cat)},
394 : std::vector<size_t>{static_cast<size_t>(number_weather_cat)}};
395 : if (emit_transformed_parameters__) {}
396 : if (emit_generated_quantities__) {}
397 : }
398 : inline void
399 : constrained_param_names(std::vector<std::string>& param_names__, bool
400 : emit_transformed_parameters__ = true, bool
401 : emit_generated_quantities__ = true) const final {
402 : for (int sym1__ = 1; sym1__ <= number_weather_cat; ++sym1__) {
403 : param_names__.emplace_back(std::string() + "a" + '.' +
404 : std::to_string(sym1__));
405 : }
406 : for (int sym1__ = 1; sym1__ <= number_weather_cat; ++sym1__) {
407 : param_names__.emplace_back(std::string() + "l" + '.' +
408 : std::to_string(sym1__));
409 : }
410 : if (emit_transformed_parameters__) {}
411 : if (emit_generated_quantities__) {}
412 : }
413 : inline void
414 : unconstrained_param_names(std::vector<std::string>& param_names__, bool
415 : emit_transformed_parameters__ = true, bool
416 : emit_generated_quantities__ = true) const final {
417 : for (int sym1__ = 1; sym1__ <= number_weather_cat; ++sym1__) {
418 : param_names__.emplace_back(std::string() + "a" + '.' +
419 : std::to_string(sym1__));
420 : }
421 : for (int sym1__ = 1; sym1__ <= number_weather_cat; ++sym1__) {
422 : param_names__.emplace_back(std::string() + "l" + '.' +
423 : std::to_string(sym1__));
424 : }
425 : if (emit_transformed_parameters__) {}
426 : if (emit_generated_quantities__) {}
427 : }
428 : inline std::string get_constrained_sizedtypes() const {
429 : return std::string("[{\"name\":\"a\",\"type\":{\"name\":\"vector\",\"length\":" + std::to_string(number_weather_cat) + "},\"block\":\"parameters\"},{\"name\":\"l\",\"type\":{\"name\":\"vector\",\"length\":" + std::to_string(number_weather_cat) + "},\"block\":\"parameters\"}]");
430 : }
431 : inline std::string get_unconstrained_sizedtypes() const {
432 : return std::string("[{\"name\":\"a\",\"type\":{\"name\":\"vector\",\"length\":" + std::to_string(number_weather_cat) + "},\"block\":\"parameters\"},{\"name\":\"l\",\"type\":{\"name\":\"vector\",\"length\":" + std::to_string(number_weather_cat) + "},\"block\":\"parameters\"}]");
433 : }
434 : // Begin method overload boilerplate
435 : template <typename RNG> inline void
436 : write_array(RNG& base_rng, Eigen::Matrix<double,-1,1>& params_r,
437 : Eigen::Matrix<double,-1,1>& vars, const bool
438 : emit_transformed_parameters = true, const bool
439 : emit_generated_quantities = true, std::ostream*
440 : pstream = nullptr) const {
441 : const size_t num_params__ = (number_weather_cat + number_weather_cat);
442 : const size_t num_transformed = emit_transformed_parameters * (0);
443 : const size_t num_gen_quantities = emit_generated_quantities * (0);
444 : const size_t num_to_write = num_params__ + num_transformed +
445 : num_gen_quantities;
446 : std::vector<int> params_i;
447 : vars = Eigen::Matrix<double,-1,1>::Constant(num_to_write,
448 : std::numeric_limits<double>::quiet_NaN());
449 : write_array_impl(base_rng, params_r, params_i, vars,
450 : emit_transformed_parameters, emit_generated_quantities, pstream);
451 : }
452 : template <typename RNG> inline void
453 : write_array(RNG& base_rng, std::vector<double>& params_r, std::vector<int>&
454 : params_i, std::vector<double>& vars, bool
455 : emit_transformed_parameters = true, bool
456 : emit_generated_quantities = true, std::ostream*
457 : pstream = nullptr) const {
458 : const size_t num_params__ = (number_weather_cat + number_weather_cat);
459 : const size_t num_transformed = emit_transformed_parameters * (0);
460 : const size_t num_gen_quantities = emit_generated_quantities * (0);
461 : const size_t num_to_write = num_params__ + num_transformed +
462 : num_gen_quantities;
463 : vars = std::vector<double>(num_to_write,
464 : std::numeric_limits<double>::quiet_NaN());
465 : write_array_impl(base_rng, params_r, params_i, vars,
466 : emit_transformed_parameters, emit_generated_quantities, pstream);
467 : }
468 : template <bool propto__, bool jacobian__, typename T_> inline T_
469 : log_prob(Eigen::Matrix<T_,-1,1>& params_r, std::ostream* pstream = nullptr) const {
470 : Eigen::Matrix<int,-1,1> params_i;
471 : return log_prob_impl<propto__, jacobian__>(params_r, params_i, pstream);
472 : }
473 : template <bool propto__, bool jacobian__, typename T_> inline T_
474 : log_prob(std::vector<T_>& params_r, std::vector<int>& params_i,
475 : std::ostream* pstream = nullptr) const {
476 : return log_prob_impl<propto__, jacobian__>(params_r, params_i, pstream);
477 : }
478 : inline void
479 : transform_inits(const stan::io::var_context& context,
480 : Eigen::Matrix<double,-1,1>& params_r, std::ostream*
481 : pstream = nullptr) const final {
482 : std::vector<double> params_r_vec(params_r.size());
483 : std::vector<int> params_i;
484 : transform_inits(context, params_i, params_r_vec, pstream);
485 : params_r = Eigen::Map<Eigen::Matrix<double,-1,1>>(params_r_vec.data(),
486 : params_r_vec.size());
487 : }
488 : inline void
489 : transform_inits(const stan::io::var_context& context, std::vector<int>&
490 : params_i, std::vector<double>& vars, std::ostream*
491 : pstream__ = nullptr) const {
492 : vars.resize(num_params_r__);
493 : transform_inits_impl(context, vars, pstream__);
494 : }
495 : inline void
496 : unconstrain_array(const std::vector<double>& params_constrained,
497 : std::vector<double>& params_unconstrained, std::ostream*
498 : pstream = nullptr) const {
499 : const std::vector<int> params_i;
500 : params_unconstrained = std::vector<double>(num_params_r__,
501 : std::numeric_limits<double>::quiet_NaN());
502 : unconstrain_array_impl(params_constrained, params_i,
503 : params_unconstrained, pstream);
504 : }
505 : inline void
506 : unconstrain_array(const Eigen::Matrix<double,-1,1>& params_constrained,
507 : Eigen::Matrix<double,-1,1>& params_unconstrained,
508 : std::ostream* pstream = nullptr) const {
509 : const std::vector<int> params_i;
510 : params_unconstrained = Eigen::Matrix<double,-1,1>::Constant(num_params_r__,
511 : std::numeric_limits<double>::quiet_NaN());
512 : unconstrain_array_impl(params_constrained, params_i,
513 : params_unconstrained, pstream);
514 : }
515 : };
516 : }
517 : using stan_model = model18558745e4a24__namespace::model18558745e4a24_;
518 : #ifndef USING_R
519 : // Boilerplate
520 : stan::model::model_base&
521 : new_model(stan::io::var_context& data_context, unsigned int seed,
522 : std::ostream* msg_stream) {
523 : stan_model* m = new stan_model(data_context, seed, msg_stream);
524 : return *m;
525 : }
526 : stan::math::profile_map& get_stan_profile_data() {
527 : return model18558745e4a24__namespace::profiles__;
528 : }
529 : #endif
530 : #endif
531 :
532 : RCPP_MODULE(stan_fit4model18558745e4a24__mod) {
533 : class_<rstan::stan_fit<stan_model, boost::random::ecuyer1988> >(
534 : "stan_fit4model18558745e4a24_")
535 :
536 : .constructor<SEXP, SEXP, SEXP>()
537 :
538 : .method(
539 : "call_sampler",
540 : &rstan::stan_fit<stan_model, boost::random::ecuyer1988>::call_sampler)
541 : .method(
542 : "param_names",
543 : &rstan::stan_fit<stan_model, boost::random::ecuyer1988>::param_names)
544 : .method("param_names_oi",
545 : &rstan::stan_fit<stan_model,
546 : boost::random::ecuyer1988>::param_names_oi)
547 : .method("param_fnames_oi",
548 : &rstan::stan_fit<stan_model,
549 : boost::random::ecuyer1988>::param_fnames_oi)
550 : .method(
551 : "param_dims",
552 : &rstan::stan_fit<stan_model, boost::random::ecuyer1988>::param_dims)
553 : .method("param_dims_oi",
554 : &rstan::stan_fit<stan_model,
555 : boost::random::ecuyer1988>::param_dims_oi)
556 : .method("update_param_oi",
557 : &rstan::stan_fit<stan_model,
558 : boost::random::ecuyer1988>::update_param_oi)
559 : .method("param_oi_tidx",
560 : &rstan::stan_fit<stan_model,
561 : boost::random::ecuyer1988>::param_oi_tidx)
562 : .method("grad_log_prob",
563 : &rstan::stan_fit<stan_model,
564 : boost::random::ecuyer1988>::grad_log_prob)
565 : .method("log_prob",
566 : &rstan::stan_fit<stan_model, boost::random::ecuyer1988>::log_prob)
567 : .method("unconstrain_pars",
568 : &rstan::stan_fit<stan_model,
569 : boost::random::ecuyer1988>::unconstrain_pars)
570 : .method("constrain_pars",
571 : &rstan::stan_fit<stan_model,
572 : boost::random::ecuyer1988>::constrain_pars)
573 : .method(
574 : "num_pars_unconstrained",
575 : &rstan::stan_fit<stan_model,
576 : boost::random::ecuyer1988>::num_pars_unconstrained)
577 : .method(
578 : "unconstrained_param_names",
579 : &rstan::stan_fit<
580 : stan_model, boost::random::ecuyer1988>::unconstrained_param_names)
581 : .method(
582 : "constrained_param_names",
583 : &rstan::stan_fit<stan_model,
584 : boost::random::ecuyer1988>::constrained_param_names)
585 : .method("standalone_gqs",
586 : &rstan::stan_fit<stan_model,
587 : boost::random::ecuyer1988>::standalone_gqs);
588 : }
589 :
590 :
591 : // declarations
592 : extern "C" {
593 : SEXP file185584cdbff65( ) ;
594 : }
595 :
596 : // definition
597 : SEXP file185584cdbff65() {
598 : return Rcpp::wrap("anon_model");
599 : }
CHECKING DATA AND PREPROCESSING FOR MODEL 'anon_model' NOW.
COMPILING MODEL 'anon_model' NOW.
STARTING SAMPLER FOR MODEL 'anon_model' NOW.
SAMPLING FOR MODEL 'anon_model' NOW (CHAIN 1).
Chain 1:
Chain 1: Gradient evaluation took 0.001781 seconds
Chain 1: 1000 transitions using 10 leapfrog steps per transition would take 17.81 seconds.
Chain 1: Adjust your expectations accordingly!
Chain 1:
Chain 1:
Chain 1: Iteration: 1 / 20000 [ 0%] (Warmup)
Chain 1: Iteration: 2000 / 20000 [ 10%] (Warmup)
Chain 1: Iteration: 2001 / 20000 [ 10%] (Sampling)
Chain 1: Iteration: 4000 / 20000 [ 20%] (Sampling)
Chain 1: Iteration: 6000 / 20000 [ 30%] (Sampling)
Chain 1: Iteration: 8000 / 20000 [ 40%] (Sampling)
Chain 1: Iteration: 10000 / 20000 [ 50%] (Sampling)
Chain 1: Iteration: 12000 / 20000 [ 60%] (Sampling)
Chain 1: Iteration: 14000 / 20000 [ 70%] (Sampling)
Chain 1: Iteration: 16000 / 20000 [ 80%] (Sampling)
Chain 1: Iteration: 18000 / 20000 [ 90%] (Sampling)
Chain 1: Iteration: 20000 / 20000 [100%] (Sampling)
Chain 1:
Chain 1: Elapsed Time: 63.03 seconds (Warm-up)
Chain 1: 507.332 seconds (Sampling)
Chain 1: 570.362 seconds (Total)
Chain 1:
5.3. Obtaining, Visualizing, and Interpreting the Posterior Results#
posterior_bikeshareis our one-stop-shop!
We obtained a total of
approximate posterior samples to be summarized in our inferential conclusions.
You can obtain the summary() statistics for the object posterior_bikeshare. These statistics are drawn from the approximate joint posterior distribution of the parameters of interest: \(a_1\), \(a_2\), \(a_3\), \(l_1\), \(l_2\), and \(l_3\). Lower and upper bounds for the 95% credible intervals are shown in columns 2.5% and 97.5%.
Metrics se_mean (Monte Carlo standard error), n_eff (effective sample size), and Rhat are model diagnostics (to be covered later on at the end of this course).
Attention
lp__ refers to the logarithm of the posterior joint density up to a constant that does not depend on the parameters of interest (we will not use it in this course).
You can find a useful optional resource if you are more interested in knowing how Stan performs its computation.
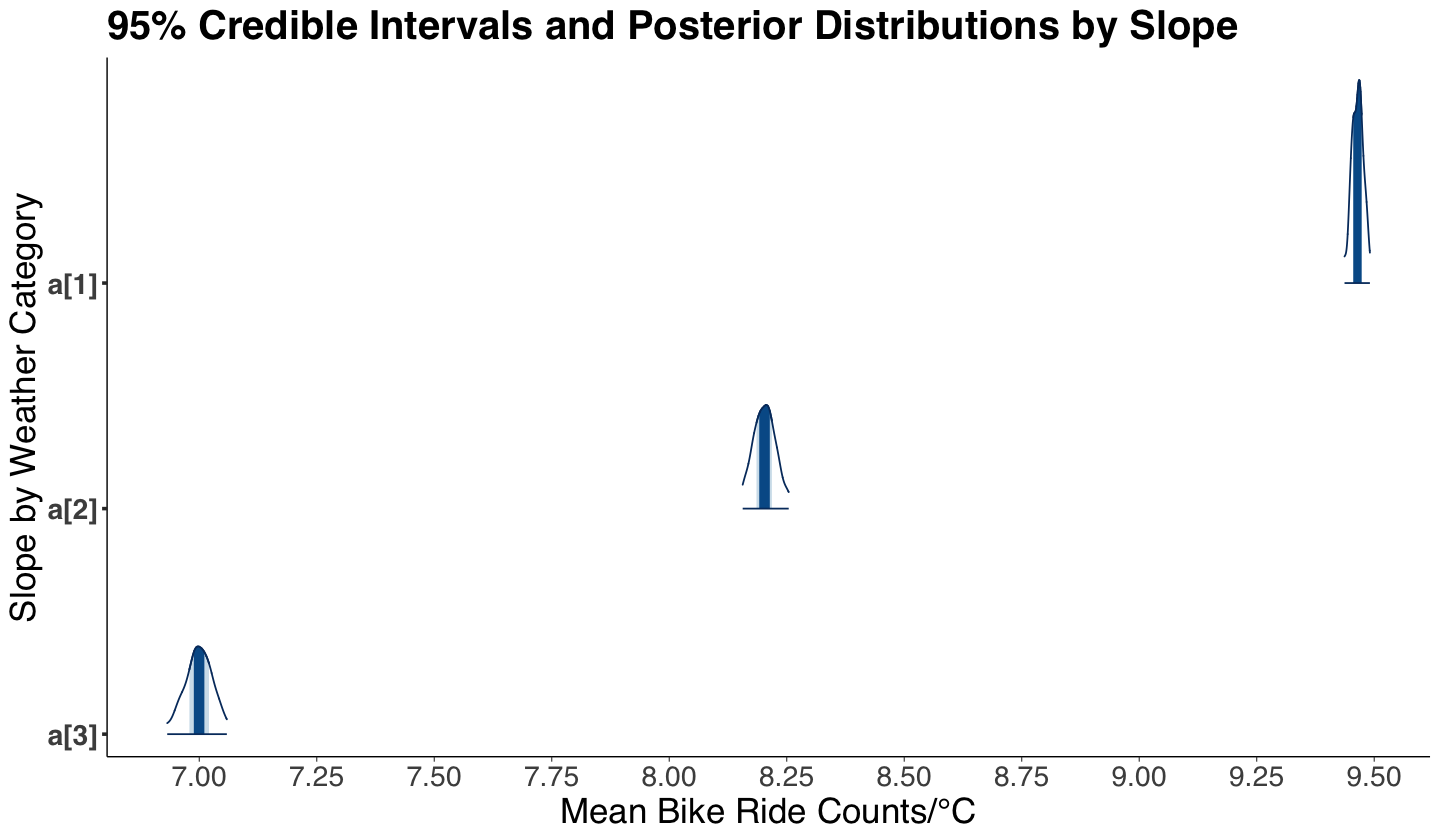
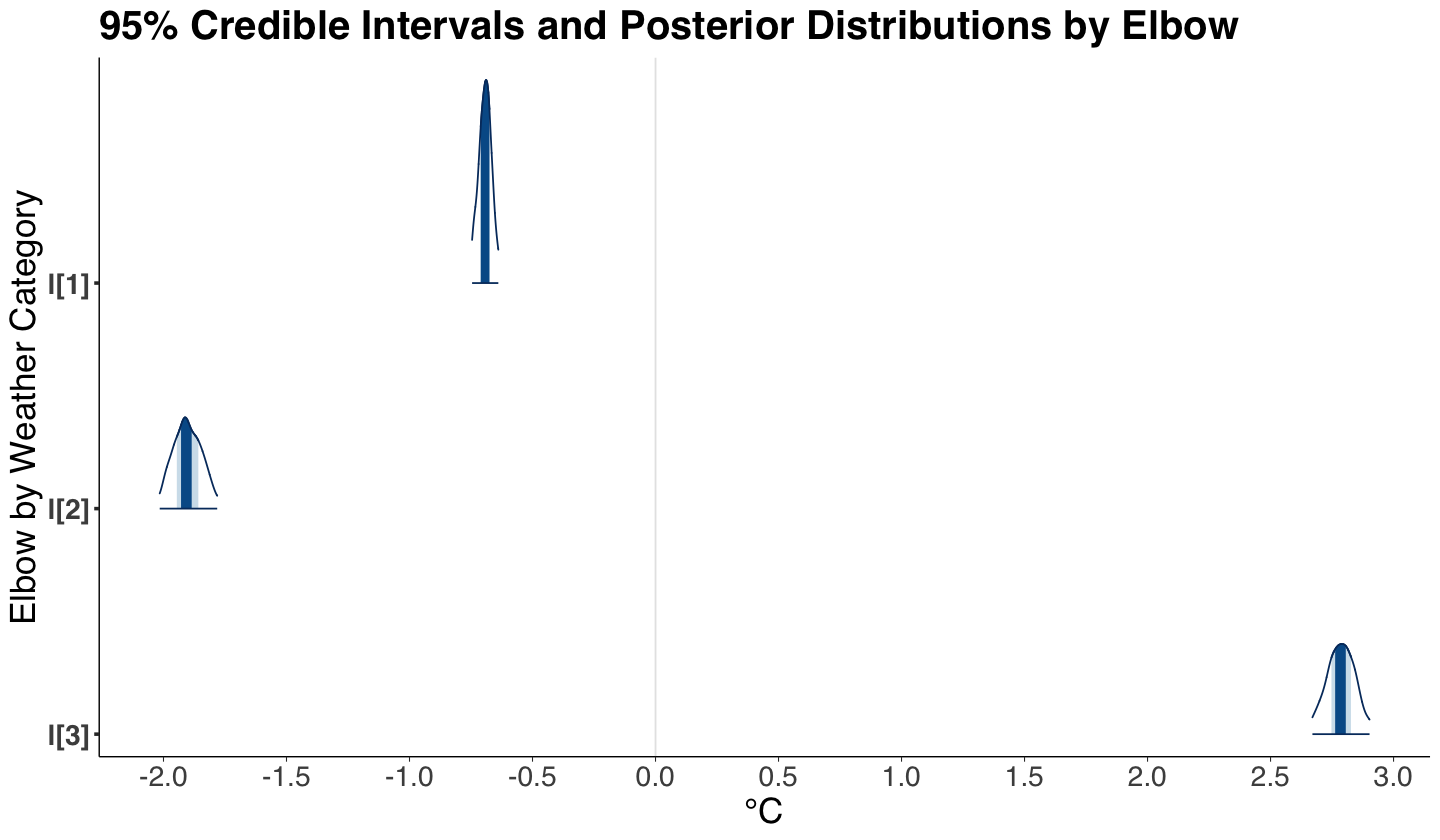
round(summary(posterior_bikeshare)$summary, 2)[-7, ]
| mean | se_mean | sd | 2.5% | 25% | 50% | 75% | 97.5% | n_eff | Rhat | |
|---|---|---|---|---|---|---|---|---|---|---|
| a[1] | 9.46 | 0 | 0.01 | 9.44 | 9.46 | 9.47 | 9.47 | 9.49 | 423.09 | 1 |
| a[2] | 8.20 | 0 | 0.03 | 8.16 | 8.19 | 8.20 | 8.22 | 8.25 | 427.82 | 1 |
| a[3] | 7.00 | 0 | 0.03 | 6.93 | 6.98 | 7.00 | 7.02 | 7.06 | 418.97 | 1 |
| l[1] | -0.69 | 0 | 0.03 | -0.75 | -0.71 | -0.69 | -0.68 | -0.64 | 454.05 | 1 |
| l[2] | -1.90 | 0 | 0.06 | -2.02 | -1.95 | -1.91 | -1.86 | -1.78 | 480.15 | 1 |
| l[3] | 2.78 | 0 | 0.06 | 2.67 | 2.75 | 2.78 | 2.83 | 2.90 | 398.77 | 1 |
Before plotting our posterior results, let us recheck the inferential inquiries:
How are temperature and weather associated with the mean number of trips?
Specifically, does the mean number of trips vary by the type of weather along with temperature?
How can we quantify this variation?
Now, we can also plot the six 95% credible intervals and posterior distributions via mcmc_areas() (from package bayesplot). Based on our summary and credible intervals, we can conclude the following (while addressing the previous inquiries):
Indeed, we have posterior evidence to state that the rates of change for mean counts of bike rides (\(a_w\)) are different by weather category (the corresponding 95% credible intervals do not overlap!). A higher rate of change mean count is associated to a nicer weather.
The temperature elbows \(l_w\) by weather category are also different (the corresponding 95% credible intervals do not overlap!).
slope_CIs <- mcmc_areas(posterior_bikeshare, pars = c("a[1]", "a[2]", "a[3]"), prob_outer = 0.95) +
theme(
plot.title = element_text(size = 24, face = "bold", family = "sans"),
axis.text.x = element_text(size = 17, angle = 0, family = "sans"),
axis.text.y = element_text(size = 17, angle = 0, family = "sans"),
axis.title = element_text(size = 21, family = "sans")
) +
ggtitle("95% Credible Intervals and Posterior Distributions by Slope") +
scale_x_continuous(breaks = seq(7, 10, 0.25)) +
labs(x = "Mean Bike Ride Counts/°C", y = "Slope by Weather Category")
slope_CIs
elbow_CIs <- mcmc_areas(posterior_bikeshare, pars = c("l[1]", "l[2]", "l[3]"), prob_outer = 0.95) +
theme(
plot.title = element_text(size = 24, face = "bold", family = "sans"),
axis.text.x = element_text(size = 17, angle = 0, family = "sans"),
axis.text.y = element_text(size = 17, angle = 0, family = "sans"),
axis.title = element_text(size = 21, family = "sans")
) +
ggtitle("95% Credible Intervals and Posterior Distributions by Elbow") +
scale_x_continuous(breaks = seq(-2.5, 3, 0.5)) +
labs(x = "°C", y = "Elbow by Weather Category")
elbow_CIs
We can obtain the posterior histograms by parameter via mcmc_hist() from bayesplot. The posterior distributions look fairly symmetric! Note that parameters \(a_w\) have positive ranges since we constrain them prior as nonnegative.
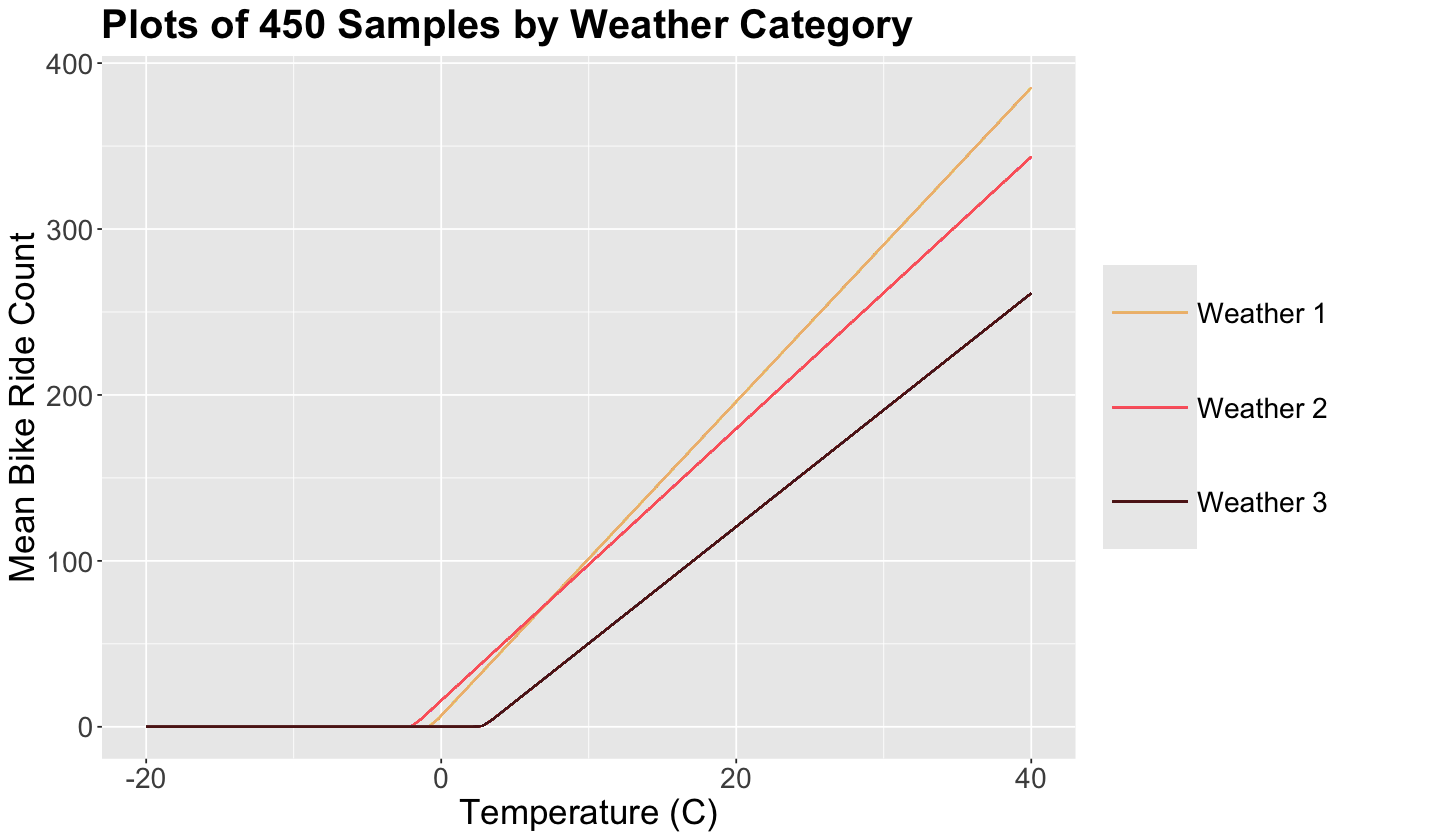
Since we have 450 sampled sets of six parameters each, we can plot 450 times our mean count equation
by weather category.
Attention
Note the low variability across our 450 samples; the lines practically overlap.
posterior_samples <- as.data.frame(posterior_bikeshare)
plot_450_samples <- ggplot() +
xlim(-20, 40) +
theme(
plot.title = element_text(size = 24, face = "bold"),
axis.text.x = element_text(size = 17, angle = 0),
axis.text.y = element_text(size = 17, angle = 0),
axis.title = element_text(size = 21),
legend.text = element_text(size = 17, margin = margin(r = 2, unit = "cm")),
legend.title = element_blank(),
legend.key.size = unit(2, 'cm')
) +
labs(y = "Mean Bike Ride Count", x = "Temperature (C)") +
ggtitle("Plots of 450 Samples by Weather Category")
# Plotting 450 lambda functions for w_1
for (i in 1:nrow(posterior_samples))
{
plot_450_samples <- plot_450_samples +
geom_function(
fun = function(x) {
log(1 + exp(posterior_samples$`a[1]`[i] *
(x - posterior_samples$`l[1]`[i])))
},
aes(colour = "Weather 1"), alpha = 0.4
)
}
# Plotting 450 lambda functions for w_2
for (i in 1:nrow(posterior_samples))
{
plot_450_samples <- plot_450_samples +
geom_function(
fun = function(x) {
log(1 + exp(posterior_samples$`a[2]`[i] *
(x - posterior_samples$`l[2]`[i])))
},
aes(colour = "Weather 2"), alpha = 0.4
)
}
# Plotting 450 lambda functions for w_3
for (i in 1:nrow(posterior_samples))
{
plot_450_samples <- plot_450_samples +
geom_function(
fun = function(x) {
log(1 + exp(posterior_samples$`a[3]`[i] *
(x - posterior_samples$`l[3]`[i])))
},
aes(colour = "Weather 3"), alpha = 0.4
)
}
The harshest weather 3 has the smallest slope and largest elbow. On the other hand, the nicest weather 1 has the largest slope but not the smallest elbow!
Exercise 14
Once we have estimated our Bayesian model, what do you think is the major drawback? Thus, how can we improve this model?
6. Wrapping Up#
We have built a Bayesian generative model totally from scratch by setting up key mathematical relations between our variables of interest.
Knowing the suitable class of distributions to use is of particular importance.
There is more than one possible modelling approach, and all models can be improved!