
Lecture 3: \(k\)-Nearest Neighbours and SVM RBFs#
UBC Master of Data Science program, 2023-24
Instructor: Varada Kolhatkar
If two things are similar, the thought of one will tend to trigger the thought of the other
– Aristotle
Lecture plan#
Announcements
Recap: Lecture 3 iClicker questions (~15 mins)
iClicker questions (~10 mins)
Class demo (~20 mins)
Break (~5 mins)
Worksheet section (~20 mins)
Intro to scaling (~5 mins)
Imports, announcements, and LOs#
Imports#
import sys
import os
import IPython
import matplotlib.pyplot as plt
import numpy as np
import pandas as pd
from IPython.display import HTML
sys.path.append(os.path.join(os.path.abspath("."), "code"))
import ipywidgets as widgets
import mglearn
from IPython.display import display
from ipywidgets import interact, interactive
from plotting_functions import *
from sklearn.dummy import DummyClassifier
from sklearn.model_selection import cross_validate, train_test_split
from utils import *
%matplotlib inline
pd.set_option("display.max_colwidth", 200)
import warnings
warnings.filterwarnings("ignore")
Learning outcomes#
From this lecture, you will be able to
explain the notion of similarity-based algorithms;
broadly describe how \(k\)-NNs use distances;
discuss the effect of using a small/large value of the hyperparameter \(k\) when using the \(k\)-NN algorithm;
describe the problem of curse of dimensionality;
explain the general idea of SVMs with RBF kernel;
broadly describe the relation of
gammaandChyperparameters of SVMs with the fundamental tradeoff.
Quick recap#
Why do we split the data?
What are the 4 types of data splits we discussed in the last lecture?
What are the benefits of cross-validation?
What is overfitting?
What’s the fundamental trade-off in supervised machine learning?
What is the golden rule of machine learning?
Important
If you want to run this notebook you will have to install ipywidgets.
Follow the installation instructions here.
Motivation and distances [video]#
Analogy-based models#
Suppose you are given the following training examples with corresponding labels and are asked to label a given test example.

An intuitive way to classify the test example is by finding the most “similar” example(s) from the training set and using that label for the test example.
Analogy-based algorithms in practice#
Herta’s High-tech Facial Recognition
Feature vectors for human faces
\(k\)-NN to identify which face is on their watch list
Recommendation systems
General idea of \(k\)-nearest neighbours algorithm#
Consider the following toy dataset with two classes.
blue circles \(\rightarrow\) class 0
red triangles \(\rightarrow\) class 1
green stars \(\rightarrow\) test examples
X, y = mglearn.datasets.make_forge()
X_test = np.array([[8.2, 3.66214339], [9.9, 3.2], [11.2, 0.5]])
plot_train_test_points(X, y, X_test)
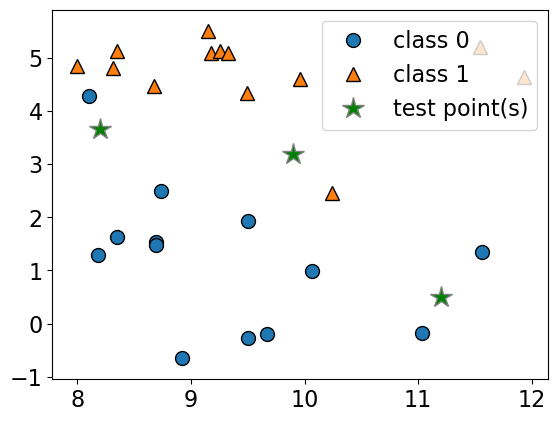
Given a new data point, predict the class of the data point by finding the “closest” data point in the training set, i.e., by finding its “nearest neighbour” or majority vote of nearest neighbours.
import panel as pn
from panel import widgets
from panel.interact import interact
import matplotlib
pn.extension()
def f(n_neighbors):
plt.clf()
fig = plt.figure(figsize=(6, 4))
plot_knn_clf(X, y, X_test, n_neighbors=n_neighbors)
return fig
n_neighbors_selector = pn.widgets.IntSlider(name='n_neighbors', start=1, end=10, value=1)
# interact(f, n_neighbors=n_neighbors_selector)
interact(f, n_neighbors=n_neighbors_selector).embed(max_opts=10)
n_neighbors 1
<Figure size 640x480 with 0 Axes>
n_neighbors 10
<Figure size 640x480 with 0 Axes>
n_neighbors 9
<Figure size 640x480 with 0 Axes>
n_neighbors
8
<Figure size 640x480 with 0 Axes>
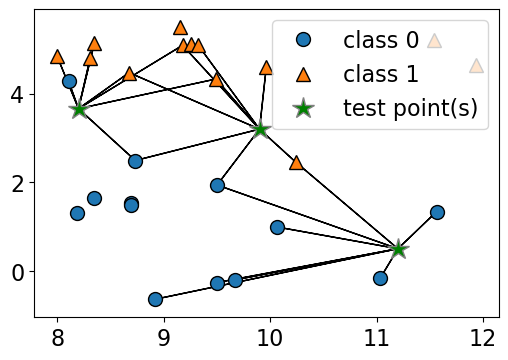
n_neighbors 7
<Figure size 640x480 with 0 Axes>
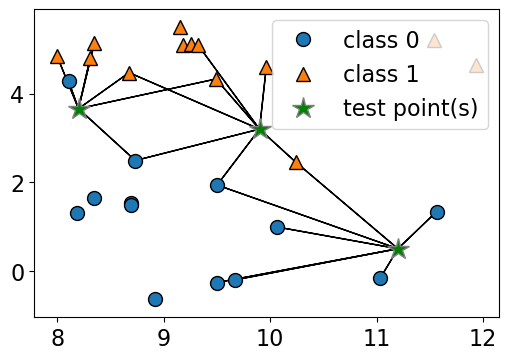
n_neighbors 6
<Figure size 640x480 with 0 Axes>
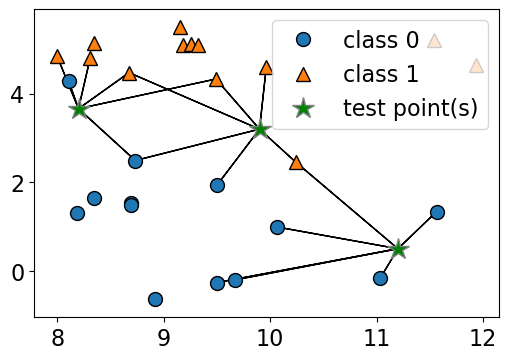
n_neighbors 5
<Figure size 640x480 with 0 Axes>
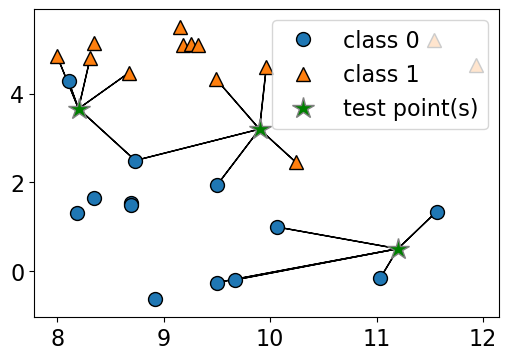
n_neighbors 4
<Figure size 640x480 with 0 Axes>
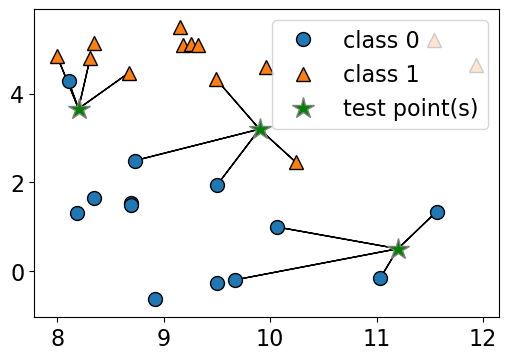
n_neighbors 3
<Figure size 640x480 with 0 Axes>
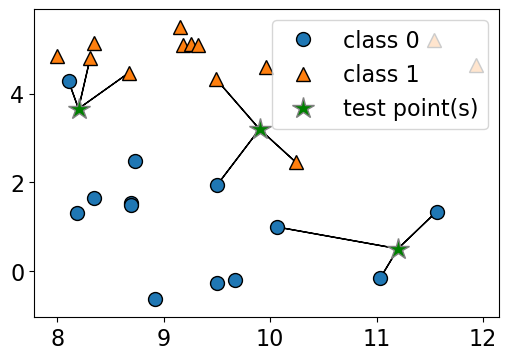
n_neighbors 2
<Figure size 640x480 with 0 Axes>
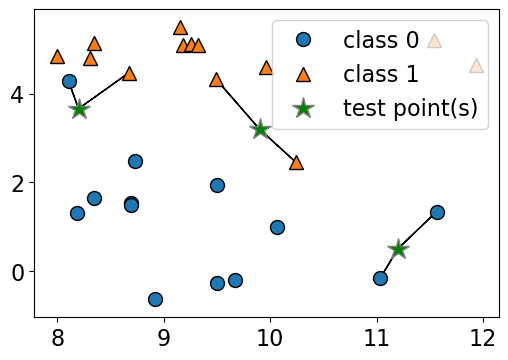
n_neighbors 1
<Figure size 640x480 with 0 Axes>

# interactive(
# f,
# n_neighbors=widgets.IntSlider(min=1, max=10, step=2, value=1),
# )
Geometric view of tabular data and dimensions#
To understand analogy-based algorithms it’s useful to think of data as points in a high dimensional space.
Our
Xrepresents the problem in terms of relevant features (\(d\)) with one dimension for each feature (column).Examples are points in a \(d\)-dimensional space.
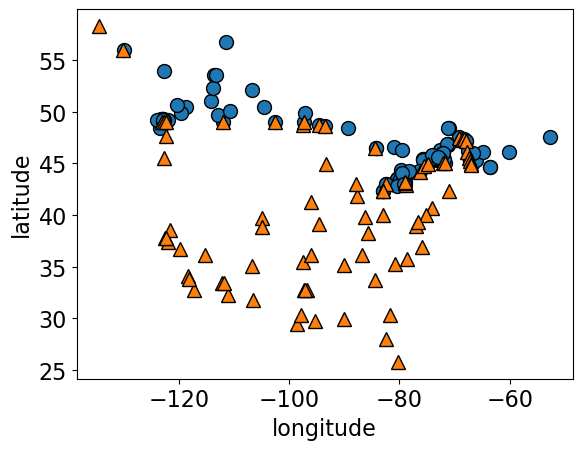
How many dimensions (features) are there in the cities data?
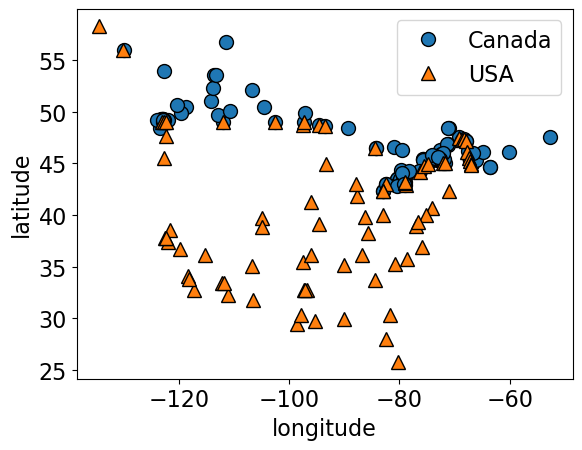
cities_df = pd.read_csv("data/canada_usa_cities.csv")
X_cities = cities_df[["longitude", "latitude"]]
y_cities = cities_df["country"]
mglearn.discrete_scatter(X_cities.iloc[:, 0], X_cities.iloc[:, 1], y_cities)
plt.xlabel("longitude")
plt.ylabel("latitude");
Recall the Spotify Song Attributes dataset from homework 1.
How many dimensions (features) we used in the homework?
spotify_df = pd.read_csv("data/spotify.csv", index_col=0)
X_spotify = spotify_df.drop(columns=["target", "song_title", "artist"])
print("The number of features in the Spotify dataset: %d" % X_spotify.shape[1])
X_spotify.head()
---------------------------------------------------------------------------
FileNotFoundError Traceback (most recent call last)
Cell In[9], line 1
----> 1 spotify_df = pd.read_csv("data/spotify.csv", index_col=0)
2 X_spotify = spotify_df.drop(columns=["target", "song_title", "artist"])
3 print("The number of features in the Spotify dataset: %d" % X_spotify.shape[1])
File ~/miniconda3/envs/571/lib/python3.10/site-packages/pandas/io/parsers/readers.py:948, in read_csv(filepath_or_buffer, sep, delimiter, header, names, index_col, usecols, dtype, engine, converters, true_values, false_values, skipinitialspace, skiprows, skipfooter, nrows, na_values, keep_default_na, na_filter, verbose, skip_blank_lines, parse_dates, infer_datetime_format, keep_date_col, date_parser, date_format, dayfirst, cache_dates, iterator, chunksize, compression, thousands, decimal, lineterminator, quotechar, quoting, doublequote, escapechar, comment, encoding, encoding_errors, dialect, on_bad_lines, delim_whitespace, low_memory, memory_map, float_precision, storage_options, dtype_backend)
935 kwds_defaults = _refine_defaults_read(
936 dialect,
937 delimiter,
(...)
944 dtype_backend=dtype_backend,
945 )
946 kwds.update(kwds_defaults)
--> 948 return _read(filepath_or_buffer, kwds)
File ~/miniconda3/envs/571/lib/python3.10/site-packages/pandas/io/parsers/readers.py:611, in _read(filepath_or_buffer, kwds)
608 _validate_names(kwds.get("names", None))
610 # Create the parser.
--> 611 parser = TextFileReader(filepath_or_buffer, **kwds)
613 if chunksize or iterator:
614 return parser
File ~/miniconda3/envs/571/lib/python3.10/site-packages/pandas/io/parsers/readers.py:1448, in TextFileReader.__init__(self, f, engine, **kwds)
1445 self.options["has_index_names"] = kwds["has_index_names"]
1447 self.handles: IOHandles | None = None
-> 1448 self._engine = self._make_engine(f, self.engine)
File ~/miniconda3/envs/571/lib/python3.10/site-packages/pandas/io/parsers/readers.py:1705, in TextFileReader._make_engine(self, f, engine)
1703 if "b" not in mode:
1704 mode += "b"
-> 1705 self.handles = get_handle(
1706 f,
1707 mode,
1708 encoding=self.options.get("encoding", None),
1709 compression=self.options.get("compression", None),
1710 memory_map=self.options.get("memory_map", False),
1711 is_text=is_text,
1712 errors=self.options.get("encoding_errors", "strict"),
1713 storage_options=self.options.get("storage_options", None),
1714 )
1715 assert self.handles is not None
1716 f = self.handles.handle
File ~/miniconda3/envs/571/lib/python3.10/site-packages/pandas/io/common.py:863, in get_handle(path_or_buf, mode, encoding, compression, memory_map, is_text, errors, storage_options)
858 elif isinstance(handle, str):
859 # Check whether the filename is to be opened in binary mode.
860 # Binary mode does not support 'encoding' and 'newline'.
861 if ioargs.encoding and "b" not in ioargs.mode:
862 # Encoding
--> 863 handle = open(
864 handle,
865 ioargs.mode,
866 encoding=ioargs.encoding,
867 errors=errors,
868 newline="",
869 )
870 else:
871 # Binary mode
872 handle = open(handle, ioargs.mode)
FileNotFoundError: [Errno 2] No such file or directory: 'data/spotify.csv'
Dimensions in ML problems#
In ML, usually we deal with high dimensional problems where examples are hard to visualize.
\(d \approx 20\) is considered low dimensional
\(d \approx 1000\) is considered medium dimensional
\(d \approx 100,000\) is considered high dimensional
Feature vectors#
- Feature vector
is composed of feature values associated with an example.
Some example feature vectors are shown below.
print(
"An example feature vector from the cities dataset: %s"
% (X_cities.iloc[0].to_numpy())
)
print(
"An example feature vector from the Spotify dataset: \n%s"
% (X_spotify.iloc[0].to_numpy())
)
An example feature vector from the cities dataset: [-130.0437 55.9773]
An example feature vector from the Spotify dataset:
[ 1.02000e-02 8.33000e-01 2.04600e+05 4.34000e-01 2.19000e-02
2.00000e+00 1.65000e-01 -8.79500e+00 1.00000e+00 4.31000e-01
1.50062e+02 4.00000e+00 2.86000e-01]
Similarity between examples#
Let’s take 2 points (two feature vectors) from the cities dataset.
two_cities = X_cities.sample(2, random_state=120)
two_cities
| longitude | latitude | |
|---|---|---|
| 69 | -104.8253 | 38.8340 |
| 35 | -112.0741 | 33.4484 |
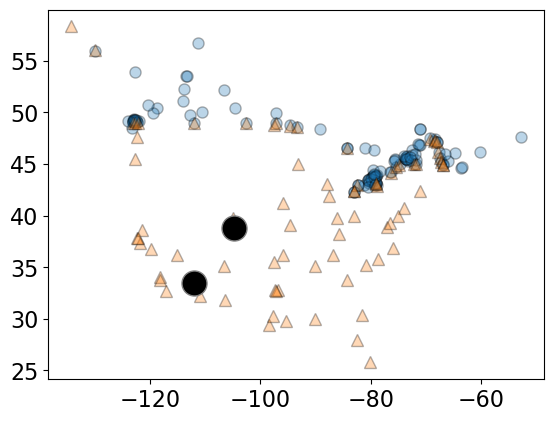
The two sampled points are shown as big black circles.
mglearn.discrete_scatter(
X_cities.iloc[:, 0], X_cities.iloc[:, 1], y_cities, s=8, alpha=0.3
)
mglearn.discrete_scatter(
two_cities.iloc[:, 0], two_cities.iloc[:, 1], markers="o", c="k", s=18
);
Distance between feature vectors#
For the cities at the two big circles, what is the distance between them?
A common way to calculate the distance between vectors is calculating the Euclidean distance.
The euclidean distance between vectors \(u = <u_1, u_2, \dots, u_n>\) and \(v = <v_1, v_2, \dots, v_n>\) is defined as:
Euclidean distance#
two_cities
| longitude | latitude | |
|---|---|---|
| 69 | -104.8253 | 38.8340 |
| 35 | -112.0741 | 33.4484 |
Subtract the two cities
Square the difference
Sum them up
Take the square root
# Subtract the two cities
print("Subtract the cities: \n%s\n" % (two_cities.iloc[1] - two_cities.iloc[0]))
# Squared sum of the difference
print(
"Sum of squares: %0.4f" % (np.sum((two_cities.iloc[1] - two_cities.iloc[0]) ** 2))
)
# Take the square root
print(
"Euclidean distance between cities: %0.4f"
% (np.sqrt(np.sum((two_cities.iloc[1] - two_cities.iloc[0]) ** 2)))
)
Subtract the cities:
longitude -7.2488
latitude -5.3856
dtype: float64
Sum of squares: 81.5498
Euclidean distance between cities: 9.0305
two_cities
| longitude | latitude | |
|---|---|---|
| 69 | -104.8253 | 38.8340 |
| 35 | -112.0741 | 33.4484 |
# Euclidean distance using sklearn
from sklearn.metrics.pairwise import euclidean_distances
euclidean_distances(two_cities)
array([[0. , 9.03049217],
[9.03049217, 0. ]])
Note: scikit-learn supports a number of other distance metrics.
Finding the nearest neighbour#
Let’s look at distances from all cities to all other cities
dists = euclidean_distances(X_cities)
np.fill_diagonal(dists, np.inf)
dists.shape
(209, 209)
pd.DataFrame(dists)
| 0 | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | ... | 199 | 200 | 201 | 202 | 203 | 204 | 205 | 206 | 207 | 208 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | inf | 4.955113 | 9.869531 | 10.106452 | 10.449666 | 19.381676 | 28.366626 | 33.283857 | 33.572105 | 36.180388 | ... | 9.834455 | 58.807684 | 16.925593 | 56.951696 | 59.384127 | 58.289799 | 64.183423 | 52.426410 | 58.033459 | 51.498562 |
| 1 | 4.955113 | inf | 14.677579 | 14.935802 | 15.305346 | 24.308448 | 33.200978 | 38.082949 | 38.359992 | 40.957919 | ... | 14.668787 | 63.533498 | 21.656349 | 61.691640 | 64.045304 | 63.032656 | 68.887343 | 57.253724 | 62.771969 | 56.252160 |
| 2 | 9.869531 | 14.677579 | inf | 0.334411 | 0.808958 | 11.115406 | 20.528403 | 25.525757 | 25.873103 | 28.479109 | ... | 0.277381 | 51.076798 | 10.783789 | 49.169693 | 51.934205 | 50.483751 | 56.512897 | 44.235152 | 50.249720 | 43.699224 |
| 3 | 10.106452 | 14.935802 | 0.334411 | inf | 0.474552 | 10.781004 | 20.194002 | 25.191396 | 25.538702 | 28.144750 | ... | 0.275352 | 50.743133 | 10.480249 | 48.836189 | 51.599860 | 50.150395 | 56.179123 | 43.904226 | 49.916254 | 43.365623 |
| 4 | 10.449666 | 15.305346 | 0.808958 | 0.474552 | inf | 10.306500 | 19.719500 | 24.716985 | 25.064200 | 27.670344 | ... | 0.675814 | 50.269880 | 10.051472 | 48.363192 | 51.125476 | 49.677629 | 55.705696 | 43.435186 | 49.443317 | 42.892477 |
| ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... |
| 204 | 58.289799 | 63.032656 | 50.483751 | 50.150395 | 49.677629 | 39.405415 | 30.043890 | 25.057003 | 24.746328 | 22.127878 | ... | 50.333340 | 0.873356 | 41.380643 | 1.345136 | 3.373031 | inf | 6.102435 | 6.957987 | 0.316363 | 6.800190 |
| 205 | 64.183423 | 68.887343 | 56.512897 | 56.179123 | 55.705696 | 45.418031 | 36.031385 | 31.032874 | 30.709185 | 28.088948 | ... | 56.358333 | 5.442806 | 47.259286 | 7.369875 | 5.108681 | 6.102435 | inf | 12.950733 | 6.303916 | 12.819584 |
| 206 | 52.426410 | 57.253724 | 44.235152 | 43.904226 | 43.435186 | 33.258427 | 24.059863 | 19.187663 | 18.932124 | 16.380495 | ... | 44.100248 | 7.767852 | 35.637982 | 5.930561 | 9.731583 | 6.957987 | 12.950733 | inf | 6.837848 | 3.322755 |
| 207 | 58.033459 | 62.771969 | 50.249720 | 49.916254 | 49.443317 | 39.167214 | 29.799983 | 24.810368 | 24.497386 | 21.878183 | ... | 50.098326 | 0.930123 | 41.121628 | 1.082749 | 3.286821 | 0.316363 | 6.303916 | 6.837848 | inf | 6.555740 |
| 208 | 51.498562 | 56.252160 | 43.699224 | 43.365623 | 42.892477 | 32.612755 | 23.244592 | 18.256813 | 17.946783 | 15.328953 | ... | 43.546610 | 7.378764 | 34.596810 | 5.473691 | 8.568009 | 6.800190 | 12.819584 | 3.322755 | 6.555740 | inf |
209 rows × 209 columns
Let’s look at the distances between City 0 and some other cities.
print("Feature vector for city 0: \n%s\n" % (X_cities.iloc[0]))
print("Distances from city 0 to the first 5 cities: %s" % (dists[0][:5]))
# We can find the closest city with `np.argmin`:
print(
"The closest city from city 0 is: %d \n\nwith feature vector: \n%s"
% (np.argmin(dists[0]), X_cities.iloc[np.argmin(dists[0])])
)
Feature vector for city 0:
longitude -130.0437
latitude 55.9773
Name: 0, dtype: float64
Distances from city 0 to the first 5 cities: [ inf 4.95511263 9.869531 10.10645223 10.44966612]
The closest city from city 0 is: 81
with feature vector:
longitude -129.9912
latitude 55.9383
Name: 81, dtype: float64
Ok, so the closest city to City 0 is City 81.
Question#
Why did we set the diagonal entries to infinity before finding the closest city?
Finding the distances to a query point#
We can also find the distances to a new “test” or “query” city:
# Let's find a city that's closest to the a query city
query_point = [[-80, 25]]
dists = euclidean_distances(X_cities, query_point)
dists[0:10]
array([[58.85545875],
[63.80062924],
[49.30530902],
[49.01473536],
[48.60495488],
[39.96834506],
[32.92852376],
[29.53520104],
[29.52881619],
[27.84679073]])
# The query point is closest to
print(
"The query point %s is closest to the city with index %d and the distance between them is: %0.4f"
% (query_point, np.argmin(dists), dists[np.argmin(dists)])
)
The query point [[-80, 25]] is closest to the city with index 72 and the distance between them is: 0.7982
\(k\)-Nearest Neighbours (\(k\)-NNs) [video]#
small_cities = cities_df.sample(30, random_state=90)
one_city = small_cities.sample(1, random_state=44)
small_train_df = pd.concat([small_cities, one_city]).drop_duplicates(keep=False)
X_small_cities = small_train_df.drop(columns=["country"]).to_numpy()
y_small_cities = small_train_df["country"].to_numpy()
test_point = one_city[["longitude", "latitude"]].to_numpy()
plot_train_test_points(
X_small_cities,
y_small_cities,
test_point,
class_names=["Canada", "USA"],
test_format="circle",
)
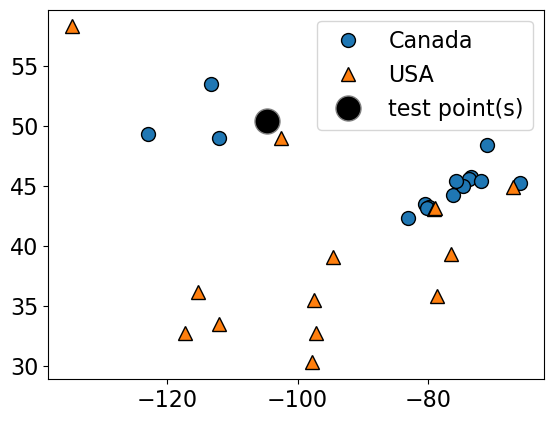
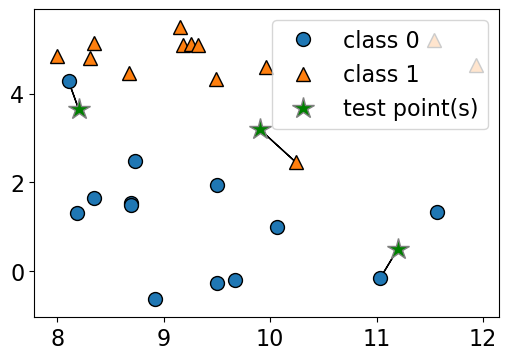
Given a new data point, predict the class of the data point by finding the “closest” data point in the training set, i.e., by finding its “nearest neighbour” or majority vote of nearest neighbours.
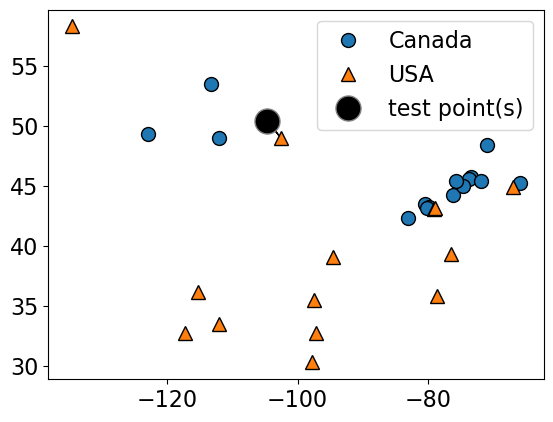
Suppose we want to predict the class of the black point.
An intuitive way to do this is predict the same label as the “closest” point (\(k = 1\)) (1-nearest neighbour)
We would predict a target of USA in this case.
plot_knn_clf(
X_small_cities,
y_small_cities,
test_point,
n_neighbors=1,
class_names=["Canada", "USA"],
test_format="circle",
)
n_neighbors 1
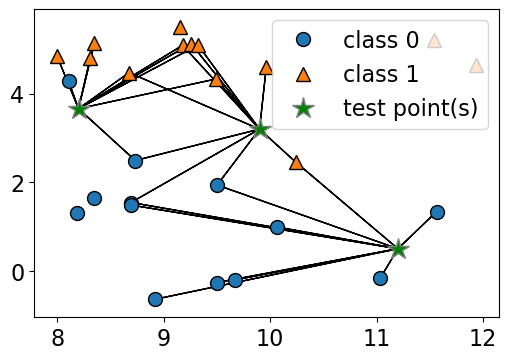
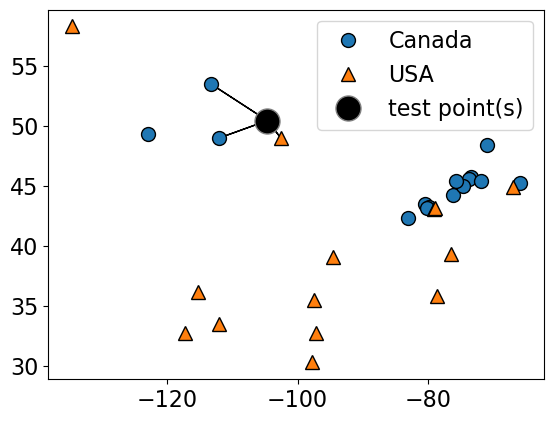
How about using \(k > 1\) to get a more robust estimate?
For example, we could also use the 3 closest points (k = 3) and let them vote on the correct class.
The Canada class would win in this case.
plot_knn_clf(
X_small_cities,
y_small_cities,
test_point,
n_neighbors=3,
class_names=["Canada", "USA"],
test_format="circle",
)
n_neighbors 3
from sklearn.neighbors import KNeighborsClassifier
k_values = [1, 3]
for k in k_values:
neigh = KNeighborsClassifier(n_neighbors=k)
neigh.fit(X_small_cities, y_small_cities)
print(
"Prediction of the black dot with %d neighbours: %s"
% (k, neigh.predict(test_point))
)
Prediction of the black dot with 1 neighbours: ['USA']
Prediction of the black dot with 3 neighbours: ['Canada']
Choosing n_neighbors#
The primary hyperparameter of the model is
n_neighbors(\(k\)) which decides how many neighbours should vote during prediction?What happens when we play around with
n_neighbors?Are we more likely to overfit with a low
n_neighborsor a highn_neighbors?Let’s examine the effect of the hyperparameter on our cities data.
X = cities_df.drop(columns=["country"])
y = cities_df["country"]
# split into train and test sets
X_train, X_test, y_train, y_test = train_test_split(
X, y, test_size=0.1, random_state=123
)
k = 1
knn1 = KNeighborsClassifier(n_neighbors=k)
scores = cross_validate(knn1, X_train, y_train, return_train_score=True)
pd.DataFrame(scores)
| fit_time | score_time | test_score | train_score | |
|---|---|---|---|---|
| 0 | 0.001642 | 0.003307 | 0.710526 | 1.0 |
| 1 | 0.001039 | 0.001988 | 0.684211 | 1.0 |
| 2 | 0.000949 | 0.001893 | 0.842105 | 1.0 |
| 3 | 0.000971 | 0.001885 | 0.702703 | 1.0 |
| 4 | 0.000897 | 0.001851 | 0.837838 | 1.0 |
k = 100
knn100 = KNeighborsClassifier(n_neighbors=k)
scores = cross_validate(knn100, X_train, y_train, return_train_score=True)
pd.DataFrame(scores)
| fit_time | score_time | test_score | train_score | |
|---|---|---|---|---|
| 0 | 0.000994 | 0.054542 | 0.605263 | 0.600000 |
| 1 | 0.001111 | 0.002233 | 0.605263 | 0.600000 |
| 2 | 0.000808 | 0.002116 | 0.605263 | 0.600000 |
| 3 | 0.000854 | 0.002164 | 0.594595 | 0.602649 |
| 4 | 0.000793 | 0.002085 | 0.594595 | 0.602649 |
def f(n_neighbors=1):
results = {}
knn = KNeighborsClassifier(n_neighbors=n_neighbors)
scores = cross_validate(knn, X_train, y_train, return_train_score=True)
results["n_neighbours"] = [n_neighbors]
results["mean_train_score"] = [round(scores["train_score"].mean(), 3)]
results["mean_valid_score"] = [round(scores["test_score"].mean(), 3)]
return(pd.DataFrame(results))
n_neighbors_selector = pn.widgets.IntSlider(name='n_neighbors', start=1, end=10, value=1)
interact(f, n_neighbors=n_neighbors_selector).embed(max_opts=20)
# interactive(
# f,
# n_neighbors=widgets.IntSlider(min=1, max=101, step=5, value=1),
# )
plot_knn_decision_boundaries(X_train, y_train, k_values=[1, 11, 100])
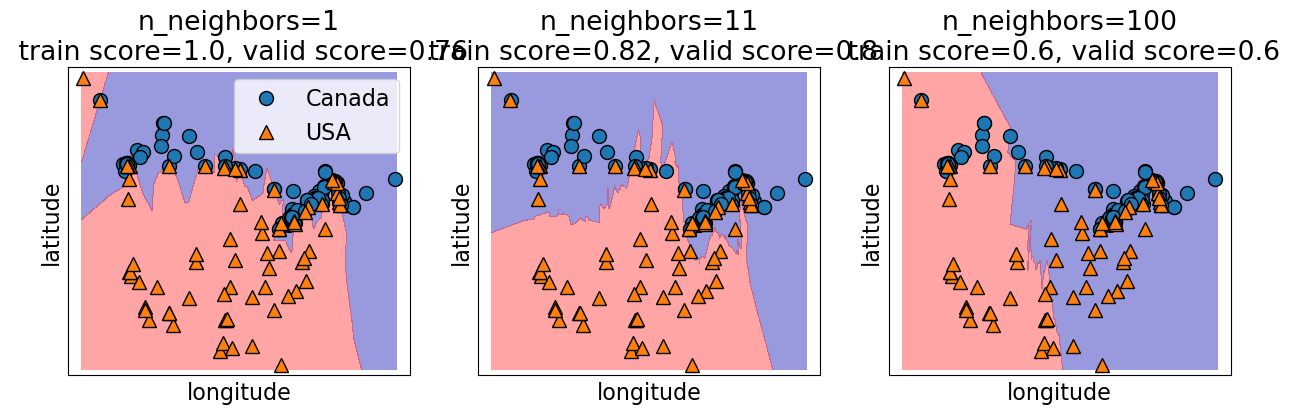
How to choose n_neighbors?#
n_neighborsis a hyperparameterWe can use hyperparameter optimization to choose
n_neighbors.
results_dict = {
"n_neighbors": [],
"mean_train_score": [],
"mean_cv_score": [],
"std_cv_score": [],
"std_train_score": [],
}
param_grid = {"n_neighbors": np.arange(1, 50, 5)}
for k in param_grid["n_neighbors"]:
knn = KNeighborsClassifier(n_neighbors=k)
scores = cross_validate(knn, X_train, y_train, return_train_score=True)
results_dict["n_neighbors"].append(k)
results_dict["mean_cv_score"].append(np.mean(scores["test_score"]))
results_dict["mean_train_score"].append(np.mean(scores["train_score"]))
results_dict["std_cv_score"].append(scores["test_score"].std())
results_dict["std_train_score"].append(scores["train_score"].std())
results_df = pd.DataFrame(results_dict)
results_df = results_df.set_index("n_neighbors")
results_df
| mean_train_score | mean_cv_score | std_cv_score | std_train_score | |
|---|---|---|---|---|
| n_neighbors | ||||
| 1 | 1.000000 | 0.755477 | 0.069530 | 0.000000 |
| 6 | 0.831135 | 0.792603 | 0.046020 | 0.013433 |
| 11 | 0.819152 | 0.802987 | 0.041129 | 0.011336 |
| 16 | 0.801863 | 0.782219 | 0.074141 | 0.008735 |
| 21 | 0.777934 | 0.766430 | 0.062792 | 0.016944 |
| 26 | 0.755364 | 0.723613 | 0.061937 | 0.025910 |
| 31 | 0.743391 | 0.707681 | 0.057646 | 0.030408 |
| 36 | 0.728777 | 0.707681 | 0.064452 | 0.021305 |
| 41 | 0.706128 | 0.681223 | 0.061241 | 0.018310 |
| 46 | 0.694155 | 0.660171 | 0.093390 | 0.018178 |
results_df[["mean_train_score", "mean_cv_score"]].plot();
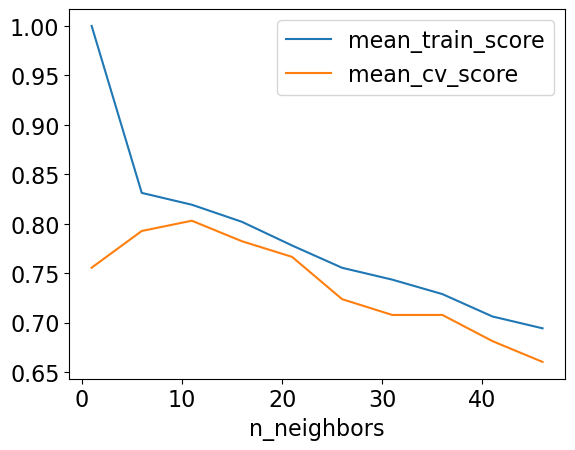
best_n_neighbours = results_df.idxmax()["mean_cv_score"]
best_n_neighbours
11
Let’s try our best model on test data.
knn = KNeighborsClassifier(n_neighbors=best_n_neighbours)
knn.fit(X_train, y_train)
print("Test accuracy: %0.3f" % (knn.score(X_test, y_test)))
Test accuracy: 0.905
Seems like we got lucky with the test set here.
❓❓ Questions for you#
(iClicker) Exercise 3.1#
iClicker cloud join link: https://join.iclicker.com/DAZZ
Select all of the following statements which are TRUE.
(A) Analogy-based models find examples from the test set that are most similar to the query example we are predicting.
(B) Euclidean distance will always have a non-negative value.
(C) With \(k\)-NN, setting the hyperparameter \(k\) to larger values typically reduces training error.
(D) Similar to decision trees, \(k\)-NNs finds a small set of good features.
(E) In \(k\)-NN, with \(k > 1\), the classification of the closest neighbour to the test example always contributes the most to the prediction.
V’s Solutions!
B
Break (5 min)#

More on \(k\)-NNs [video]#
Other useful arguments of KNeighborsClassifier#
weights\(\rightarrow\) When predicting label, you can assign higher weight to the examples which are closer to the query example.Exercise for you: Play around with this argument. Do you get a better validation score?
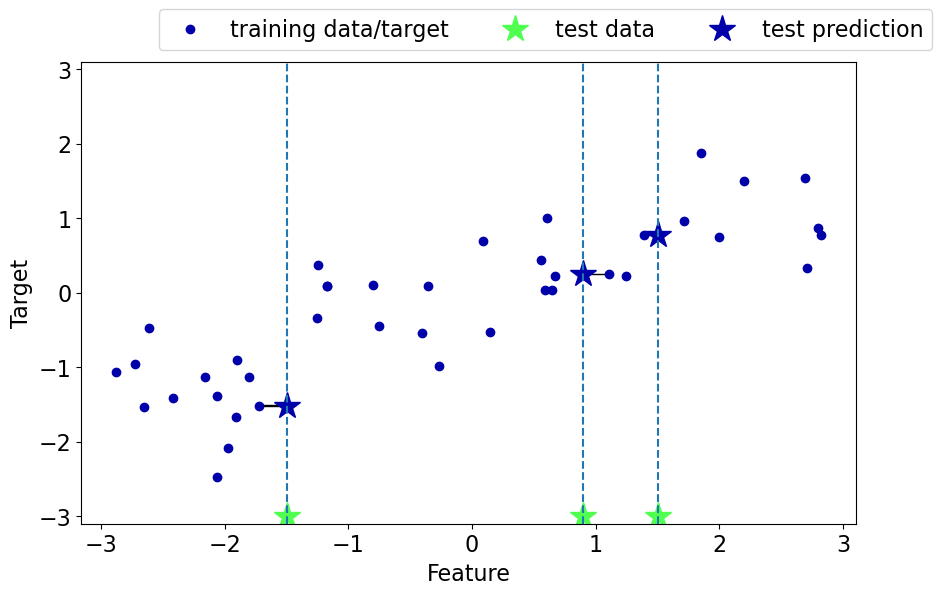
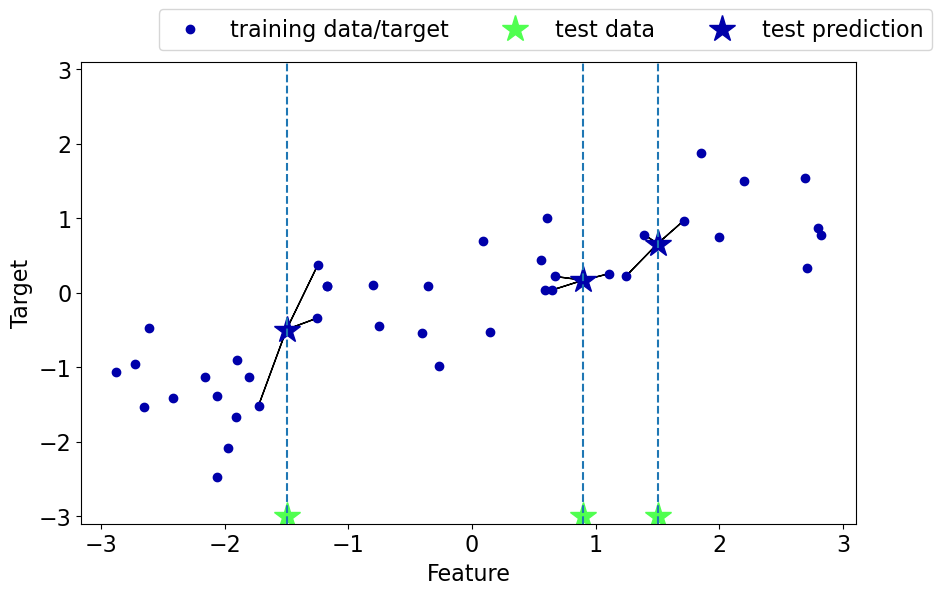
Regression with \(k\)-nearest neighbours (\(k\)-NNs)#
Can we solve regression problems with \(k\)-nearest neighbours algorithm?
In \(k\)-NN regression we take the average of the \(k\)-nearest neighbours.
We can also have weighted regression.
See an example of regression in the lecture notes.
mglearn.plots.plot_knn_regression(n_neighbors=1)
mglearn.plots.plot_knn_regression(n_neighbors=3)
Pros of \(k\)-NNs for supervised learning#
Easy to understand, interpret.
Simple hyperparameter \(k\) (
n_neighbors) controlling the fundamental tradeoff.Can learn very complex functions given enough data.
Lazy learning: Takes no time to
fit
Cons of \(k\)-NNs for supervised learning#
Can be potentially be VERY slow during prediction time, especially when the training set is very large.
Often not that great test accuracy compared to the modern approaches.
It does not work well on datasets with many features or where most feature values are 0 most of the time (sparse datasets).
Attention
For regular \(k\)-NN for supervised learning (not with sparse matrices), you should scale your features. We’ll be looking into it soon.
(Optional) Parametric vs non parametric#
You might see a lot of definitions of these terms.
A simple way to think about this is:
do you need to store at least \(O(n)\) worth of stuff to make predictions? If so, it’s non-parametric.
Non-parametric example: \(k\)-NN is a classic example of non-parametric models.
Parametric example: decision stump
If you want to know more about this, find some reading material here, here, and here.
By the way, the terms “parametric” and “non-paramteric” are often used differently by statisticians, see here for more…
Note
\(\mathcal{O}(n)\) is referred to as big \(\mathcal{O}\) notation. It tells you how fast an algorithm is or how much storage space it requires. For example, in simple terms, if you have \(n\) examples and you need to store them all you can say that the algorithm requires \(\mathcal{O}(n)\) worth of stuff.
Curse of dimensionality#
Affects all learners but especially bad for nearest-neighbour.
\(k\)-NN usually works well when the number of dimensions \(d\) is small but things fall apart quickly as \(d\) goes up.
If there are many irrelevant attributes, \(k\)-NN is hopelessly confused because all of them contribute to finding similarity between examples.
With enough irrelevant attributes the accidental similarity swamps out meaningful similarity and \(k\)-NN is no better than random guessing.
from sklearn.datasets import make_classification
nfeats_accuracy = {"nfeats": [], "dummy_valid_accuracy": [], "KNN_valid_accuracy": []}
for n_feats in range(4, 2000, 100):
X, y = make_classification(n_samples=2000, n_features=n_feats, n_classes=2)
X_train, X_test, y_train, y_test = train_test_split(
X, y, test_size=0.2, random_state=123
)
dummy = DummyClassifier(strategy="most_frequent")
dummy_scores = cross_validate(dummy, X_train, y_train, return_train_score=True)
knn = KNeighborsClassifier()
scores = cross_validate(knn, X_train, y_train, return_train_score=True)
nfeats_accuracy["nfeats"].append(n_feats)
nfeats_accuracy["KNN_valid_accuracy"].append(np.mean(scores["test_score"]))
nfeats_accuracy["dummy_valid_accuracy"].append(np.mean(dummy_scores["test_score"]))
pd.DataFrame(nfeats_accuracy)
| nfeats | dummy_valid_accuracy | KNN_valid_accuracy | |
|---|---|---|---|
| 0 | 4 | 0.508125 | 0.893750 |
| 1 | 104 | 0.503125 | 0.775000 |
| 2 | 204 | 0.510625 | 0.745000 |
| 3 | 304 | 0.508125 | 0.630625 |
| 4 | 404 | 0.499375 | 0.673750 |
| 5 | 504 | 0.500625 | 0.649375 |
| 6 | 604 | 0.502500 | 0.656875 |
| 7 | 704 | 0.511875 | 0.588125 |
| 8 | 804 | 0.506250 | 0.591250 |
| 9 | 904 | 0.502500 | 0.587500 |
| 10 | 1004 | 0.503750 | 0.615000 |
| 11 | 1104 | 0.507500 | 0.611875 |
| 12 | 1204 | 0.499375 | 0.612500 |
| 13 | 1304 | 0.501250 | 0.593750 |
| 14 | 1404 | 0.500000 | 0.550000 |
| 15 | 1504 | 0.501875 | 0.565000 |
| 16 | 1604 | 0.511250 | 0.612500 |
| 17 | 1704 | 0.511875 | 0.586875 |
| 18 | 1804 | 0.508125 | 0.603125 |
| 19 | 1904 | 0.503750 | 0.611250 |
Support Vector Machines (SVMs) with RBF kernel [video]#
Very high-level overview
Our goals here are
Use
scikit-learn’s SVM model.Broadly explain the notion of support vectors.
Broadly explain the similarities and differences between \(k\)-NNs and SVM RBFs.
Explain how
Candgammahyperparameters control the fundamental tradeoff.
(Optional) RBF stands for radial basis functions. We won’t go into what it means in this video. Refer to this video if you want to know more.
Overview#
Another popular similarity-based algorithm is Support Vector Machines with RBF Kernel (SVM RBFs)
Superficially, SVM RBFs are more like weighted \(k\)-NNs.
The decision boundary is defined by a set of positive and negative examples and their weights together with their similarity measure.
A test example is labeled positive if on average it looks more like positive examples than the negative examples.
The primary difference between \(k\)-NNs and SVM RBFs is that
Unlike \(k\)-NNs, SVM RBFs only remember the key examples (support vectors).
SVMs use a different similarity metric which is called a “kernel”. A popular kernel is Radial Basis Functions (RBFs)
They usually perform better than \(k\)-NNs!
Let’s explore SVM RBFs#
Let’s try SVMs on the cities dataset.
mglearn.discrete_scatter(X_cities.iloc[:, 0], X_cities.iloc[:, 1], y_cities)
plt.xlabel("longitude")
plt.ylabel("latitude")
plt.legend(loc=1);
X_train, X_test, y_train, y_test = train_test_split(
X_cities, y_cities, test_size=0.2, random_state=123
)
knn = KNeighborsClassifier(n_neighbors=best_n_neighbours)
scores = cross_validate(knn, X_train, y_train, return_train_score=True)
print("Mean validation score %0.3f" % (np.mean(scores["test_score"])))
pd.DataFrame(scores)
Mean validation score 0.803
| fit_time | score_time | test_score | train_score | |
|---|---|---|---|---|
| 0 | 0.002949 | 0.004925 | 0.794118 | 0.819549 |
| 1 | 0.000957 | 0.001959 | 0.764706 | 0.819549 |
| 2 | 0.000978 | 0.001963 | 0.727273 | 0.850746 |
| 3 | 0.000903 | 0.001909 | 0.787879 | 0.828358 |
| 4 | 0.000822 | 0.001737 | 0.939394 | 0.783582 |
from sklearn.svm import SVC
svm = SVC(gamma=0.01) # Ignore gamma for now
scores = cross_validate(svm, X_train, y_train, return_train_score=True)
print("Mean validation score %0.3f" % (np.mean(scores["test_score"])))
pd.DataFrame(scores)
Mean validation score 0.820
| fit_time | score_time | test_score | train_score | |
|---|---|---|---|---|
| 0 | 0.003523 | 0.001714 | 0.823529 | 0.842105 |
| 1 | 0.001296 | 0.000831 | 0.823529 | 0.842105 |
| 2 | 0.001290 | 0.000809 | 0.727273 | 0.858209 |
| 3 | 0.001253 | 0.000785 | 0.787879 | 0.843284 |
| 4 | 0.001286 | 0.000872 | 0.939394 | 0.805970 |
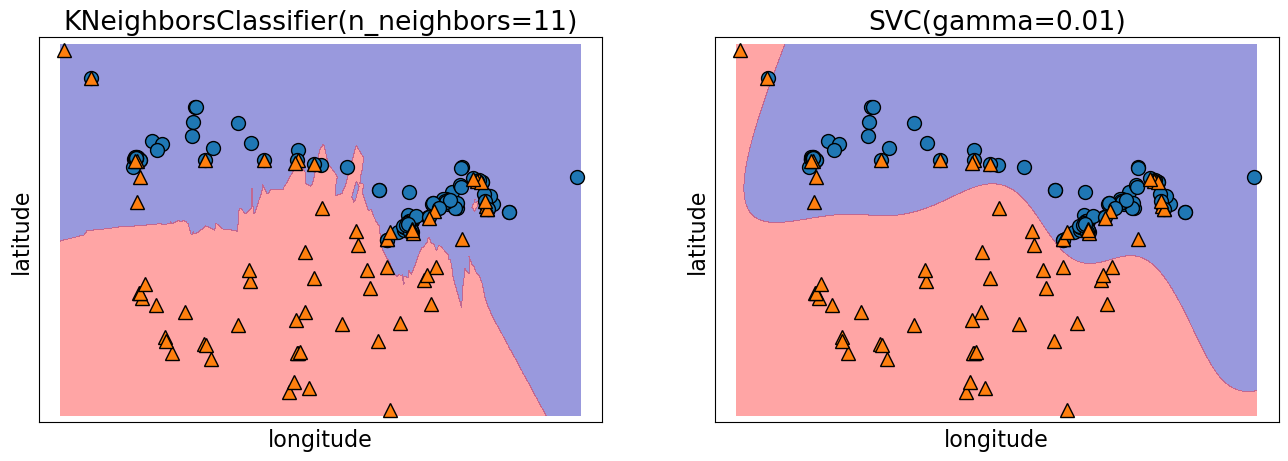
Decision boundary of SVMs#
We can think of SVM with RBF kernel as “smooth KNN”.
fig, axes = plt.subplots(1, 2, figsize=(16, 5))
for clf, ax in zip([knn, svm], axes):
clf.fit(X_train, y_train)
mglearn.plots.plot_2d_separator(
clf, X_train.to_numpy(), fill=True, eps=0.5, ax=ax, alpha=0.4
)
mglearn.discrete_scatter(X_train.iloc[:, 0], X_train.iloc[:, 1], y_train, ax=ax)
ax.set_title(clf)
ax.set_xlabel("longitude")
ax.set_ylabel("latitude")
Support vectors#
Each training example either is or isn’t a “support vector”.
This gets decided during
fit.
Main insight: the decision boundary only depends on the support vectors.
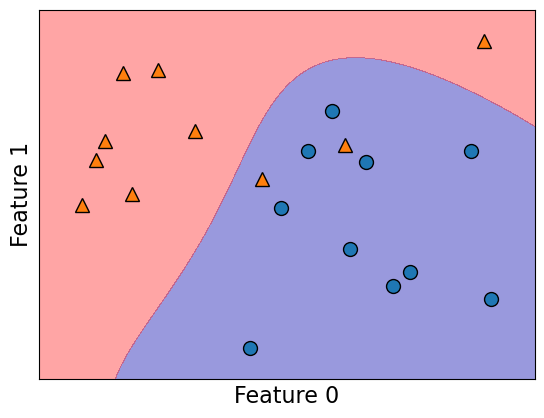
Let’s look at the support vectors.
from sklearn.datasets import make_blobs
n = 20
n_classes = 2
X_toy, y_toy = make_blobs(
n_samples=n, centers=n_classes, random_state=300
) # Let's generate some fake data
mglearn.discrete_scatter(X_toy[:, 0], X_toy[:, 1], y_toy)
plt.xlabel("Feature 0")
plt.ylabel("Feature 1")
svm = SVC(kernel="rbf", C=10, gamma=0.1).fit(X_toy, y_toy)
mglearn.plots.plot_2d_separator(svm, X_toy, fill=True, eps=0.5, alpha=0.4)
svm.support_
array([ 3, 8, 9, 14, 19, 1, 4, 6, 17], dtype=int32)
plot_support_vectors(svm, X_toy, y_toy)
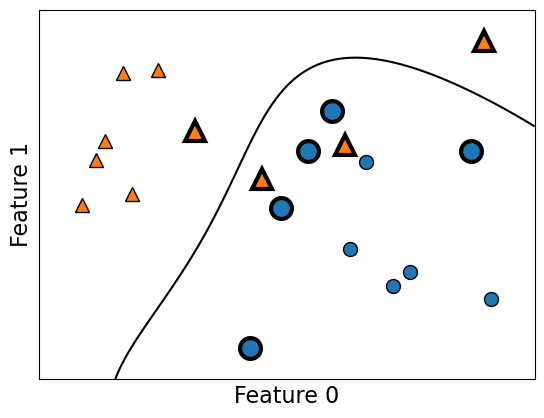
The support vectors are the bigger points in the plot above.
Hyperparameters of SVM#
Key hyperparameters of
rbfSVM aregammaC
We are not equipped to understand the meaning of these parameters at this point but you are expected to describe their relation to the fundamental tradeoff.
Relation of gamma and the fundamental trade-off#
gammacontrols the complexity (fundamental trade-off), just like other hyperparameters we’ve seen.larger
gamma\(\rightarrow\) more complexsmaller
gamma\(\rightarrow\) less complex
gamma = [0.001, 0.01, 0.1, 1.0, 10.0]
plot_svc_gamma(
gamma,
X_train.to_numpy(),
y_train.to_numpy(),
x_label="longitude",
y_label="latitude",
)
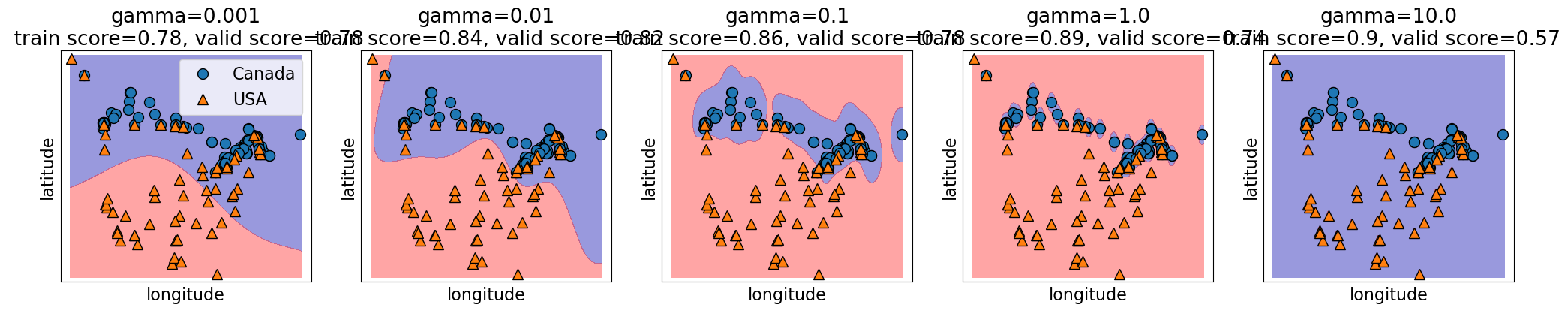
Relation of C and the fundamental trade-off#
Calso affects the fundamental tradeofflarger
C\(\rightarrow\) more complexsmaller
C\(\rightarrow\) less complex
C = [0.1, 1.0, 100.0, 1000.0, 100000.0]
plot_svc_C(
C, X_train.to_numpy(), y_train.to_numpy(), x_label="longitude", y_label="latitude"
)
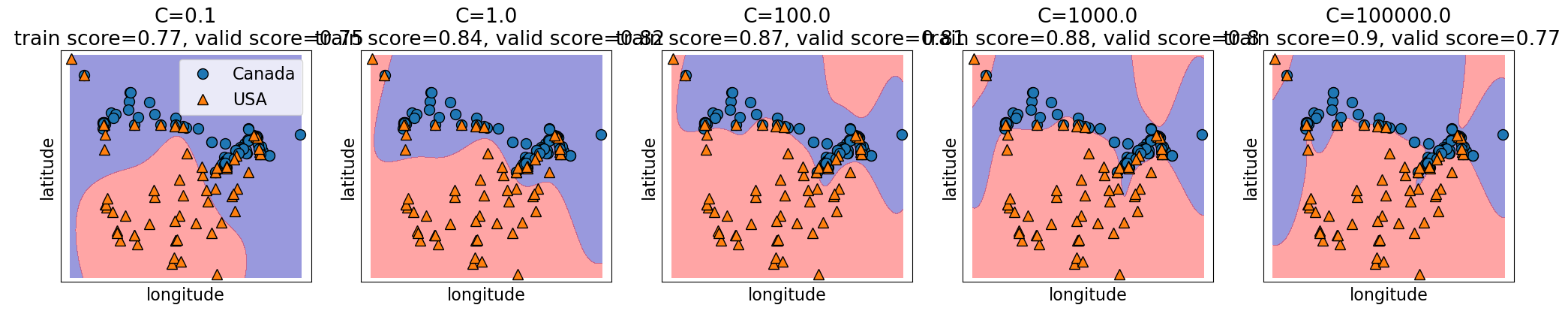
Search over multiple hyperparameters#
So far you have seen how to carry out search over a hyperparameter
In the above case the best training error is achieved by the most complex model (large
gamma, largeC).Best validation error requires a hyperparameter search to balance the fundamental tradeoff.
In general we can’t search them one at a time.
More on this next week. But if you cannot wait till then, you may look up the following:
SVM Regressor#
Similar to KNNs, you can use SVMs for regression problems as well.
See
sklearn.svm.SVRfor more details.
❓❓ Questions for you#
(iClicker) Exercise 3.2#
iClicker cloud join link: https://join.iclicker.com/DAZZ
Select all of the following statements which are TRUE.
(A) \(k\)-NN may perform poorly in high-dimensional space (say, d > 1000).
(B) In sklearn’s SVC classifier, large values of gamma tend to result in higher training score but probably lower validation score.
(C) If we increase both gamma and C, we can’t be certain if the model becomes more complex or less complex.
V’s Solutions!
A, B
More practice questions#
Check out some more practice questions here.
Summary#
We have KNNs and SVMs as new supervised learning techniques in our toolbox.
These are analogy-based learners and the idea is to assign nearby points the same label.
Unlike decision trees, all features are equally important.
Both can be used for classification or regression (much like the other methods we’ve seen).
Coming up:#
Lingering questions:
Are we ready to do machine learning on real-world datasets?
What would happen if we use \(k\)-NNs or SVM RBFs on the spotify dataset from hw1?
What happens if we have missing values in our data?
What do we do if we have features with categories or string values?

