library(ggexpress)When conducting Exploratory Data Analysis it is useful to plot the variables in the data to get an initial sense of the distribution and potential behaviour of the data. If the dataset contains many variables creating separate plots for each one could become tedious. This package automates the plot configuration process and generates basic graphics that summarize the data.
This package contains 4 functions; two for general purpose exploratory tasks and two that are more specific.
Creates a basic histogram that indicates the position of the mean and median and displays the standard deviation.
gghist
Creates a scatterplot and calculates the correlation coefficient.
ggscatter
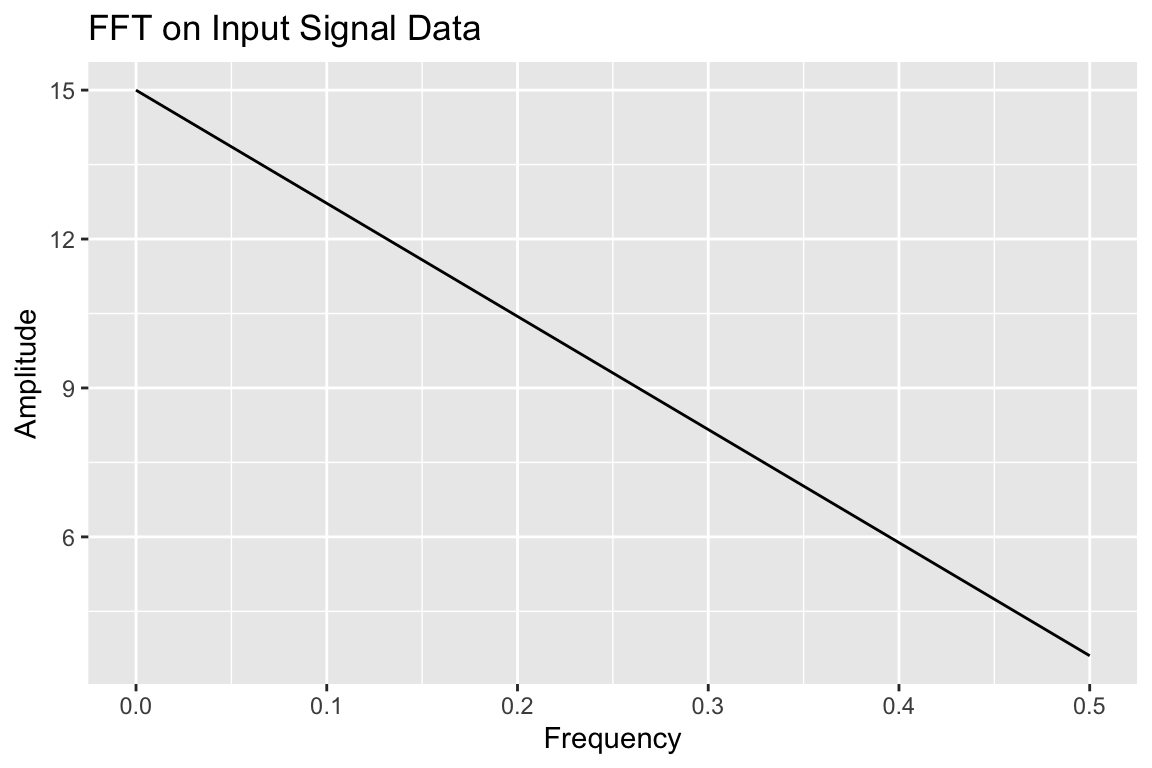
Creates a Fourier transform plot.
fourier_transform
Converts time series data into 4 subplots displaying the raw data, trend, seasonal and noise components.
ts_plot
Data: gapminder
The gapminder::gapminder dataset will be used to explore the basic functionality of each of the plots.
library(gapminder)
gapminder::gapminder
#> # A tibble: 1,704 x 6
#> country continent year lifeExp pop gdpPercap
#> <fct> <fct> <int> <dbl> <int> <dbl>
#> 1 Afghanistan Asia 1952 28.8 8425333 779.
#> 2 Afghanistan Asia 1957 30.3 9240934 821.
#> 3 Afghanistan Asia 1962 32.0 10267083 853.
#> 4 Afghanistan Asia 1967 34.0 11537966 836.
#> 5 Afghanistan Asia 1972 36.1 13079460 740.
#> 6 Afghanistan Asia 1977 38.4 14880372 786.
#> 7 Afghanistan Asia 1982 39.9 12881816 978.
#> 8 Afghanistan Asia 1987 40.8 13867957 852.
#> 9 Afghanistan Asia 1992 41.7 16317921 649.
#> 10 Afghanistan Asia 1997 41.8 22227415 635.
#> # … with 1,694 more rowsObtain a histogram and basic summary statistics with gghist()
This plot displays the position of the mean and median of life expectancy from the gapminder dataset. In addition, the plot also displays the value of the mean, median and standard deviation.
gghist(data = gapminder, variable = lifeExp)
#> `stat_bin()` using `bins = 30`. Pick better value with `binwidth`.
#> `stat_bin()` using `bins = 30`. Pick better value with `binwidth`.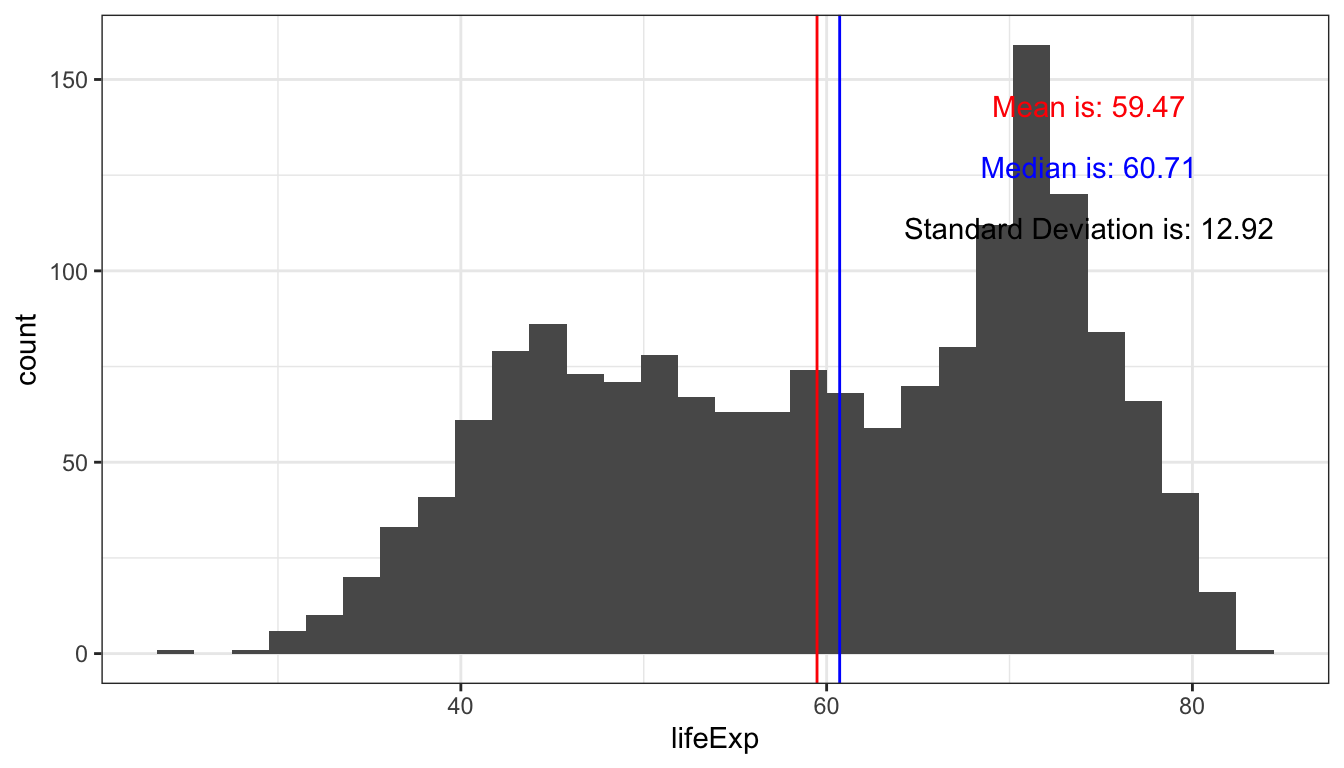
This plot shows the distribution of gdp per capita.
gghist(data = gapminder, variable = gdpPercap)
#> `stat_bin()` using `bins = 30`. Pick better value with `binwidth`.
#> `stat_bin()` using `bins = 30`. Pick better value with `binwidth`.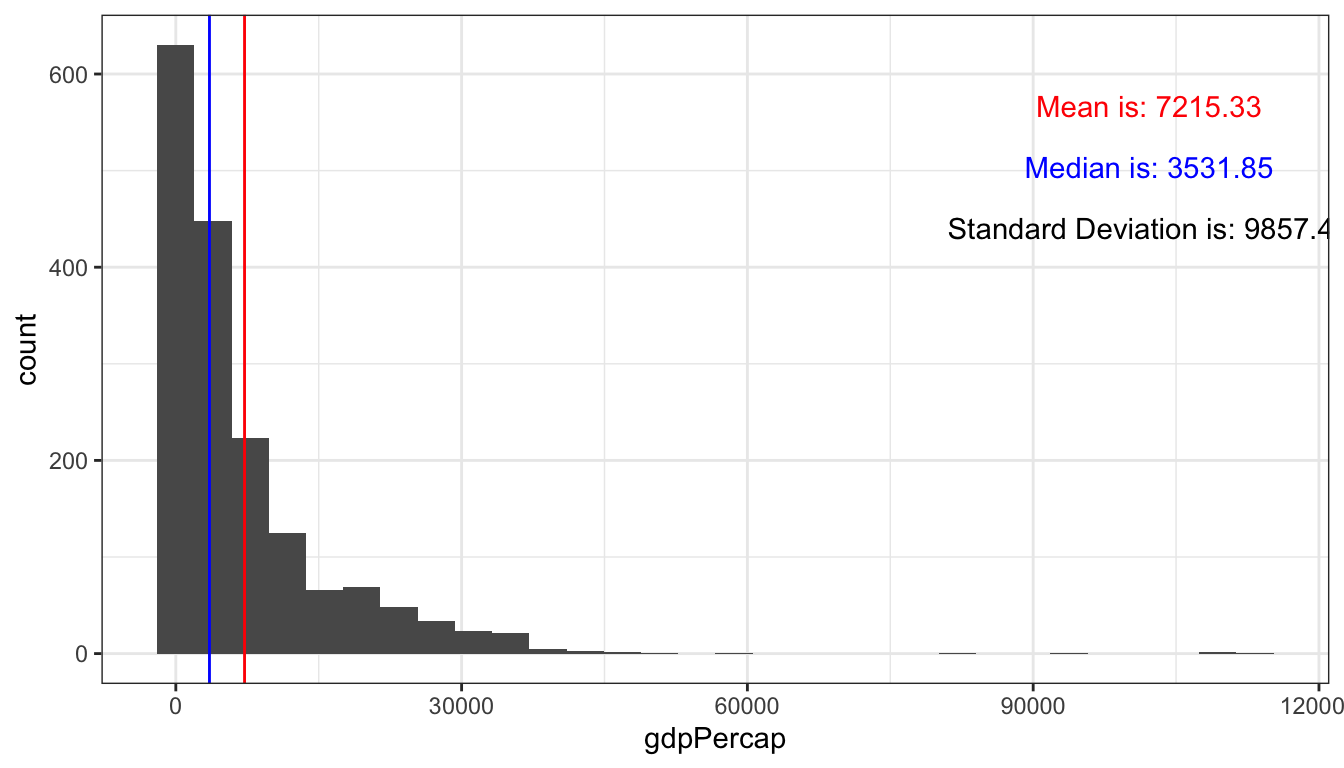
Obtain a scatterplot with scatter_express()
The
scatter_express()returns a basic scatterplot but also returns the correlation coefficient between the two variables. The plot below shows the relationship between petal length and petal width in iris flowers.
scatter_express(df = iris,
xval = Petal.Length,
yval = Petal.Width)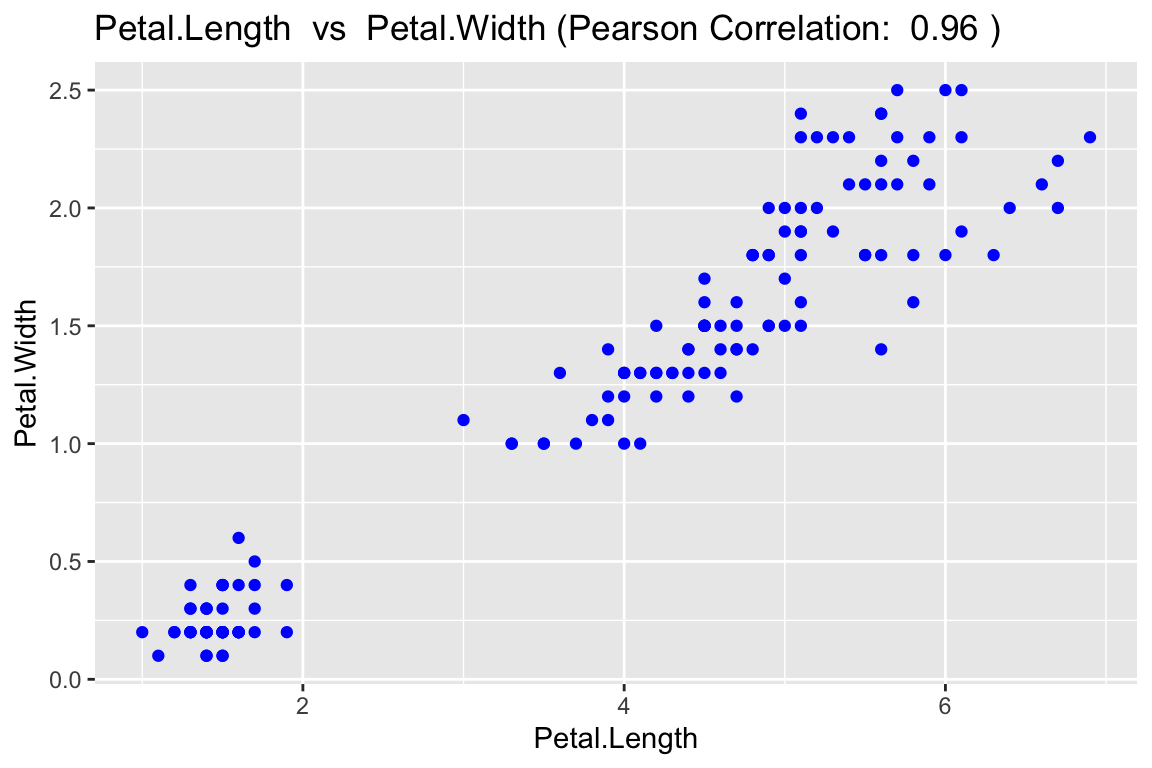
In addition, scatter_express() provides the option display the data on a log transformed scale for data with non-linear relationships. The first plot displays the GDP per capita vs the life expectancy before before log transformation.
scatter_express(df = gapminder,
xval = gdpPercap,
yval = lifeExp)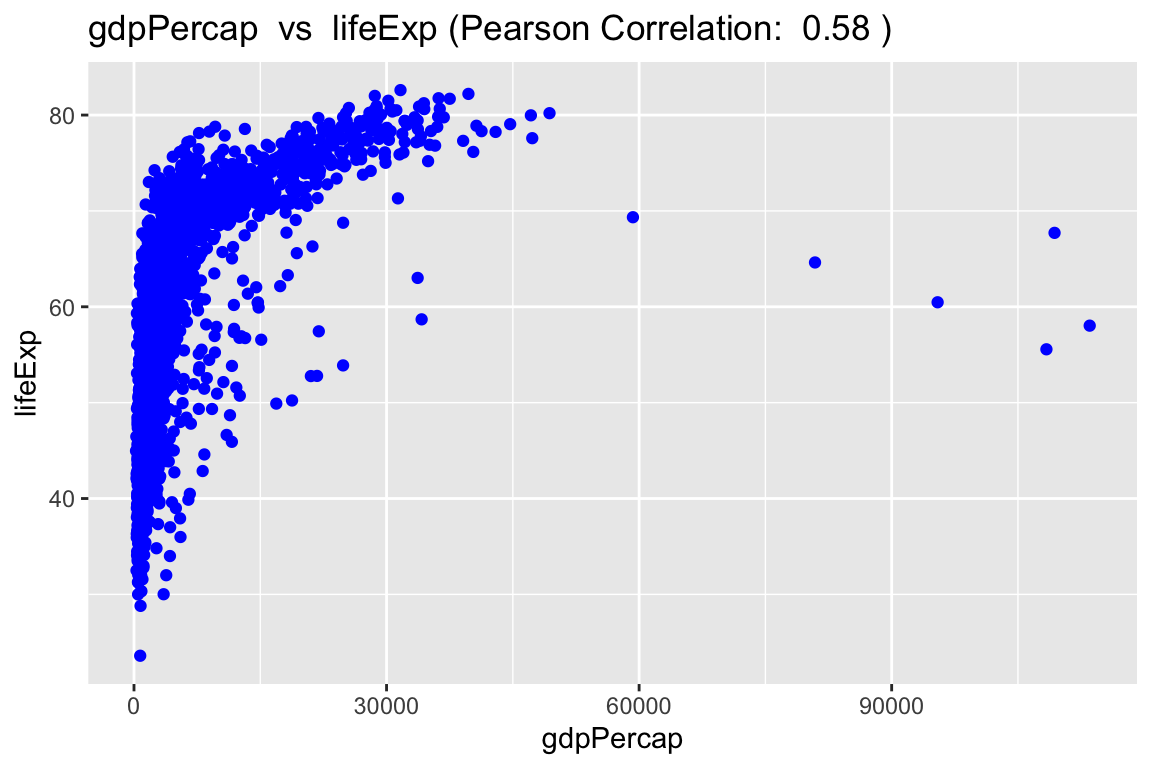
Once the log tranform is applied, a linear trend beetween GDP per capita and life expectancy becomes more apparent.
scatter_express(df = gapminder,
xval = gdpPercap,
yval = lifeExp,
x_transform = TRUE,
y_transform = TRUE)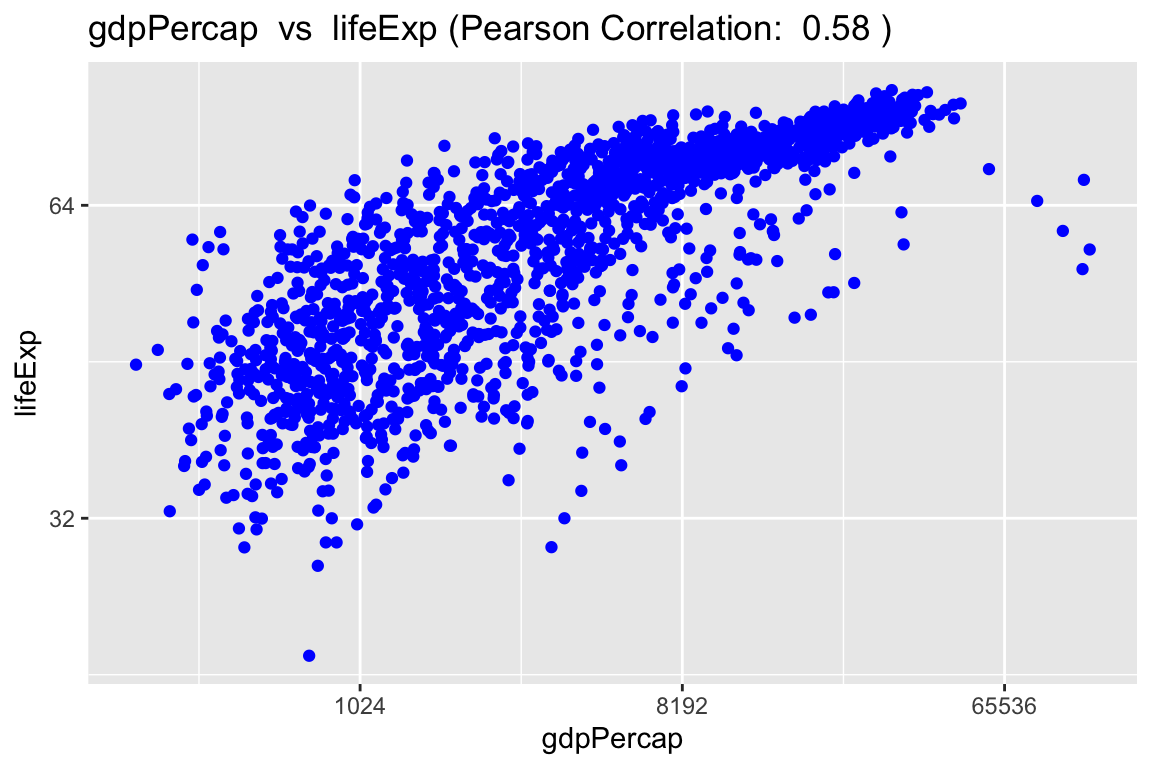
Time Series Decomposition with ts_plot()
This function, ts_plot(), is able to take in a time stamped dataframe and convert it into a time series object. The time series is then decomposed into its trend, seasonsal and white noise components.
# sample data
time <- c("1950 Q1", "1950 Q2", "1950 Q3", "1950 Q4",
"1951 Q1", "1951 Q2", "1951 Q3", "1951 Q4",
"1952 Q1", "1952 Q2", "1952 Q3", "1952 Q4")
earnings <- c(0.71, 0.63, 0.82, 0.91,
0.51, 1.31, 1.82, 2.01,
0.63, 0.82, 0.91, 0.71)
ts_data <- tibble::tibble(time, earnings)
ts_plot(ts_data, "earnings", 4)
#> Registered S3 method overwritten by 'quantmod':
#> method from
#> as.zoo.data.frame zoo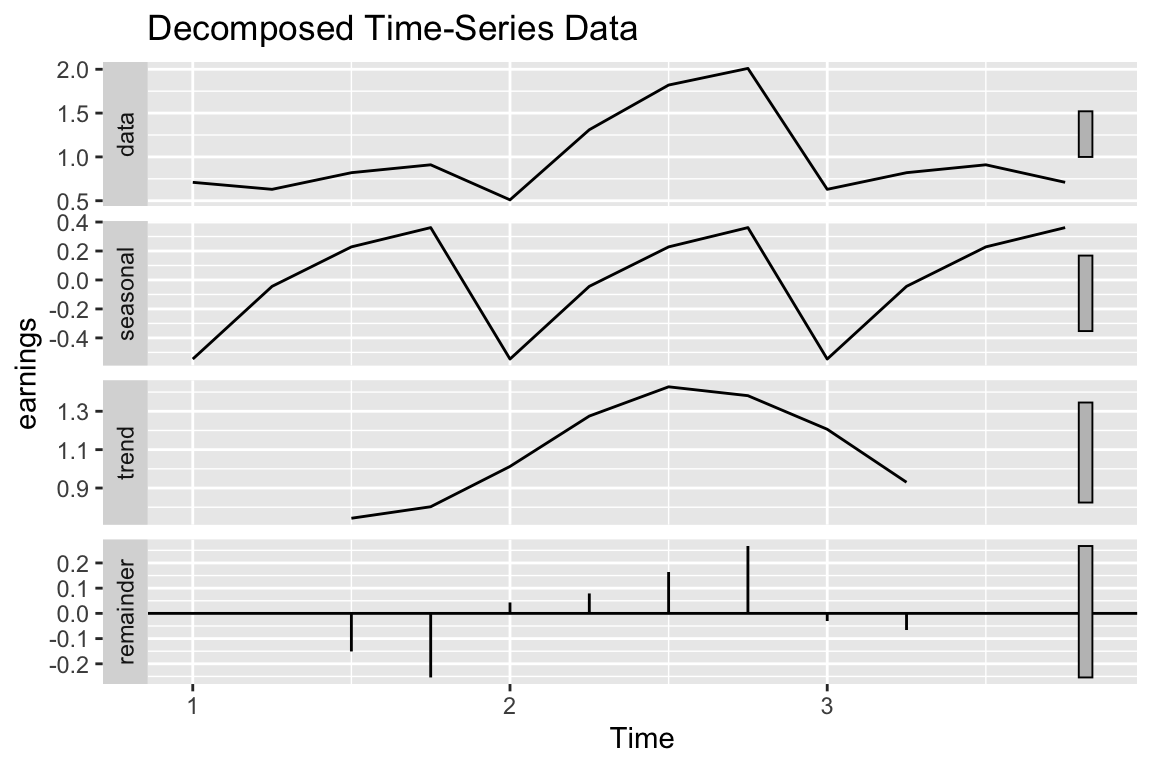
Fourier Transform plot with fourier_transform()
# sample data
my_data = tibble::tibble(time_series = c(0, 1, 2, 3), signal = c(2, 3, 4, 6))
fourier_transform(data = my_data,
time_col = "time_series",
data_col = "signal")