Connectivity Visualizations¶
Connectivity plots¶
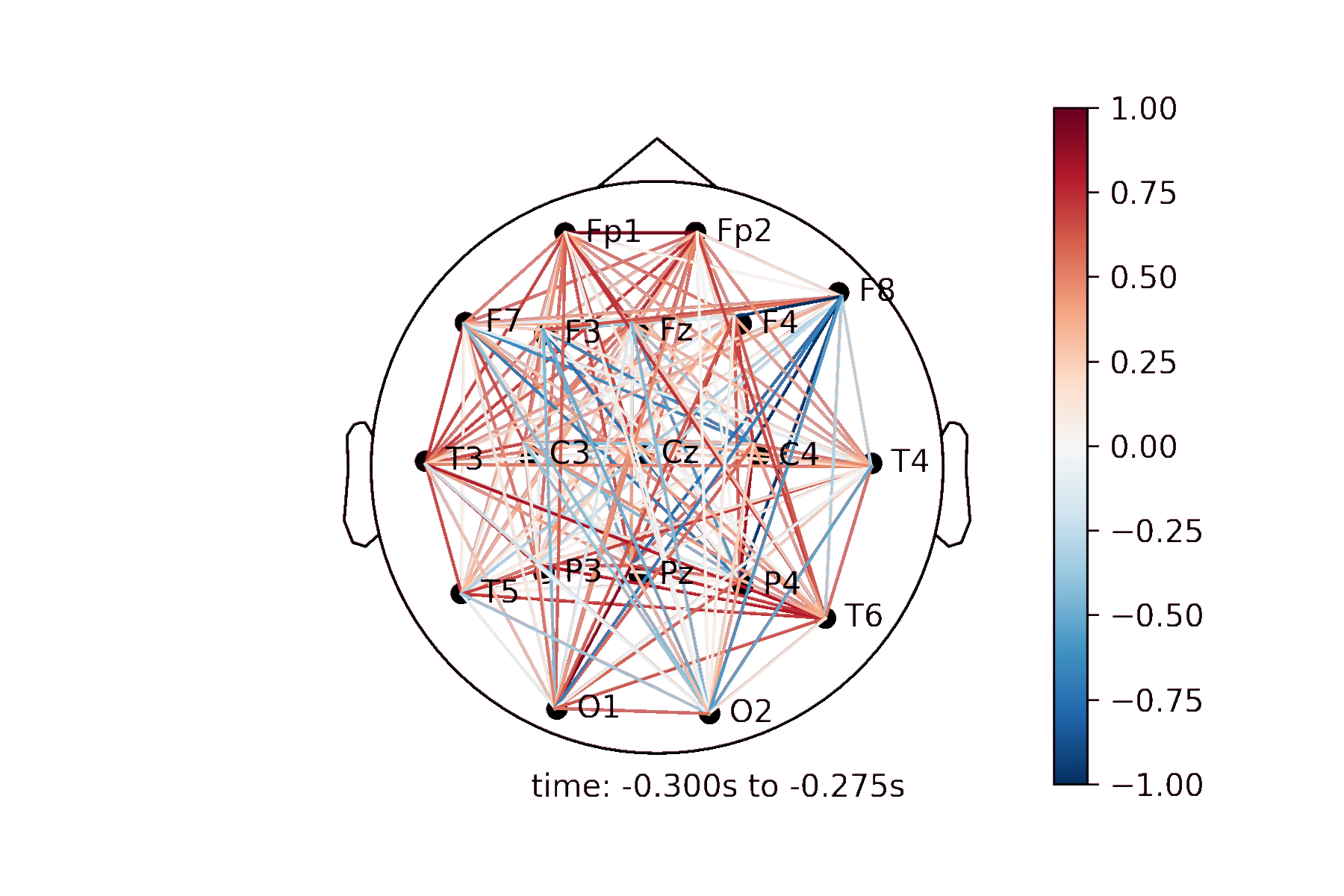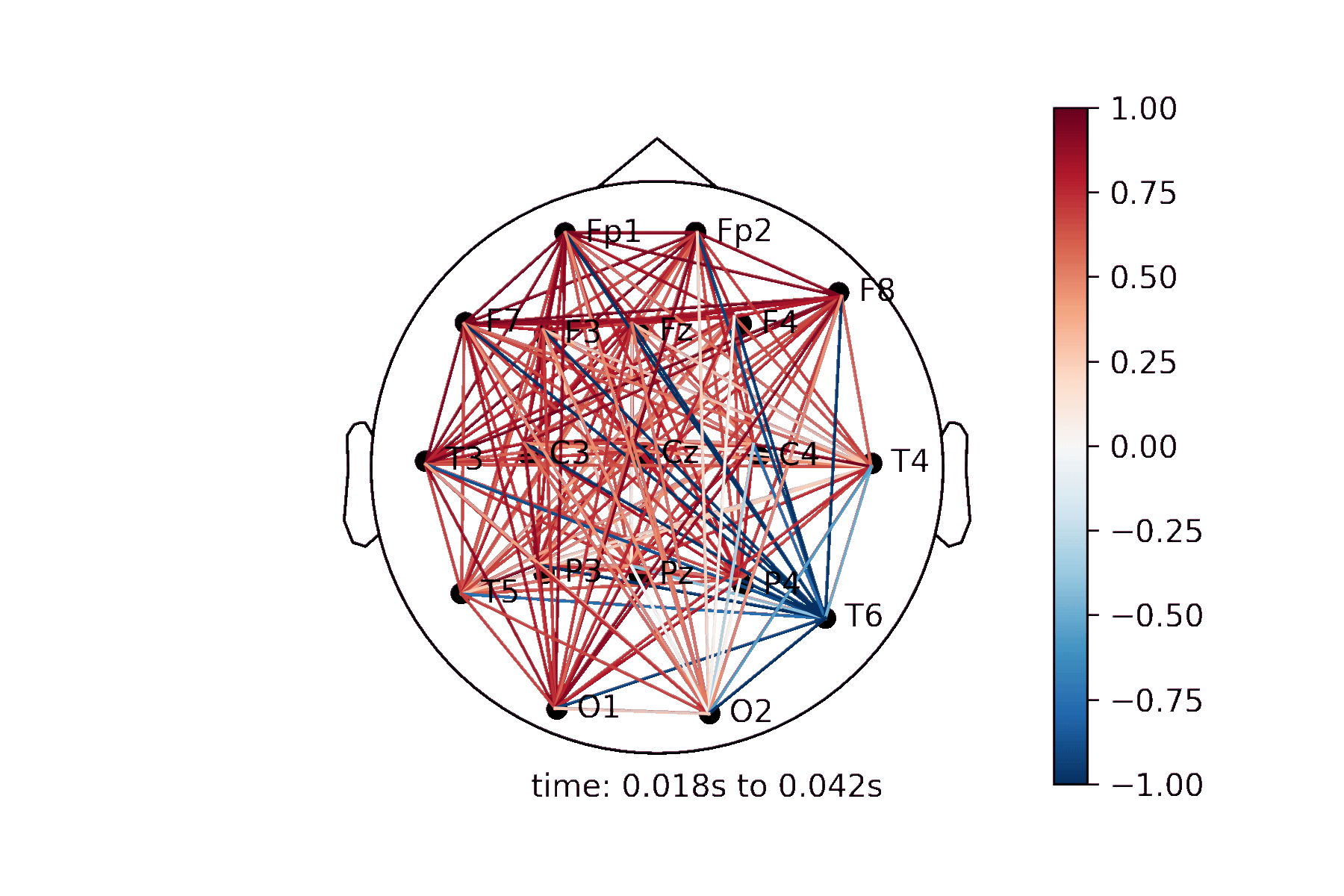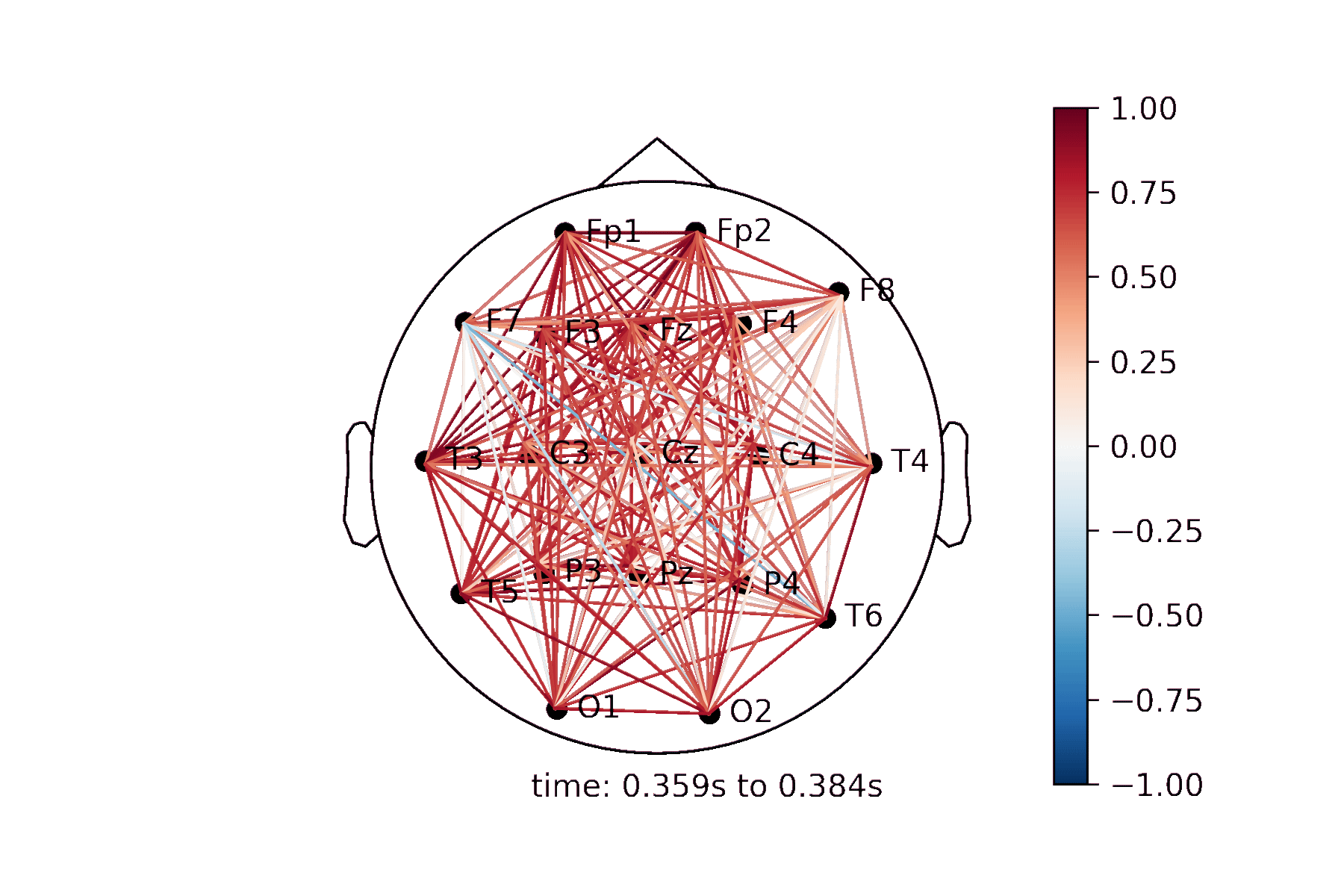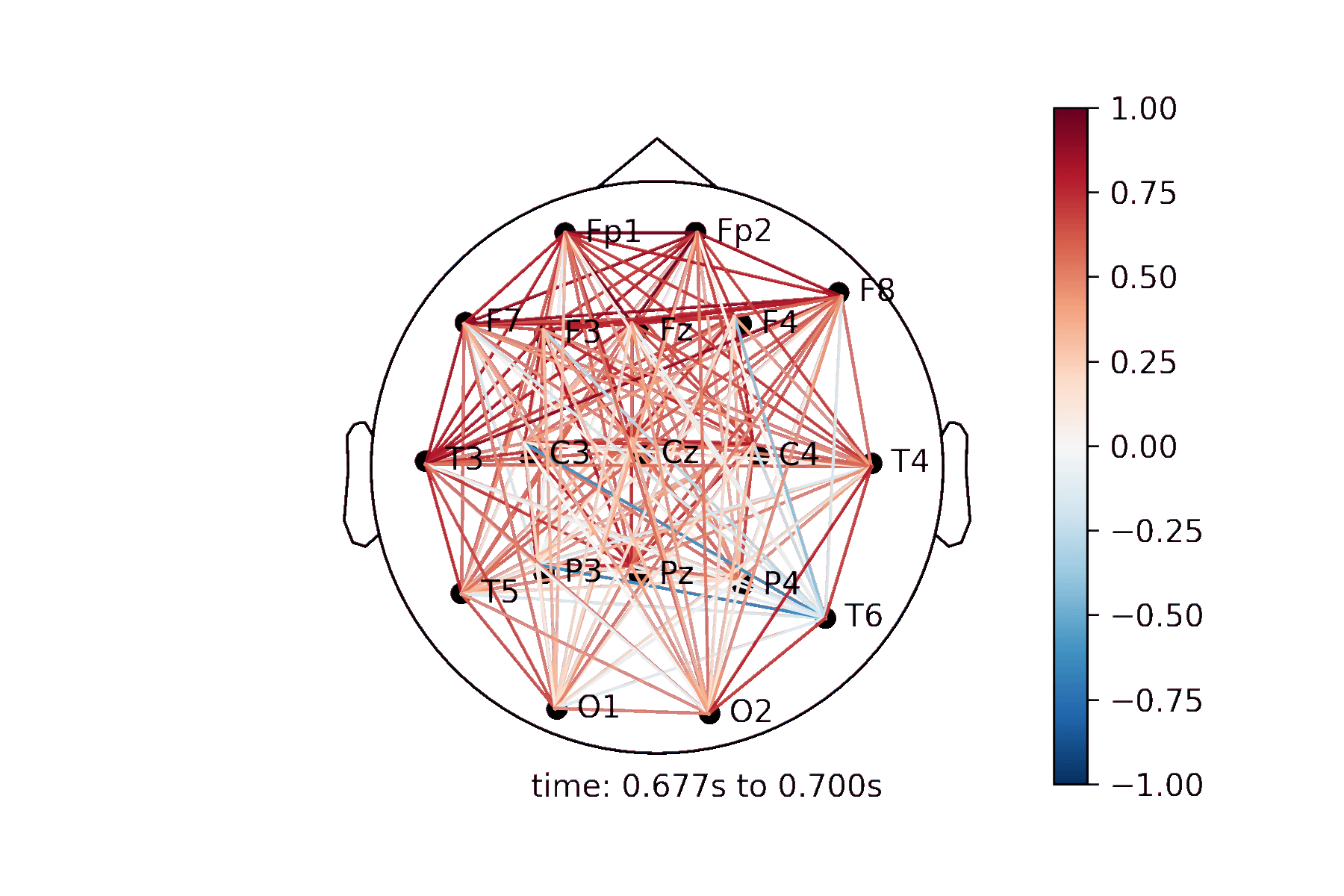
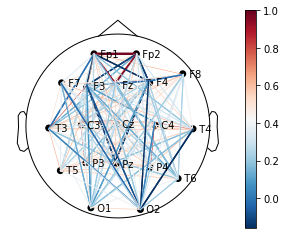
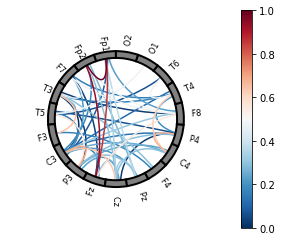
The connectivity plots provide a way to visualize pair-wise correlation, coherence, and connectivity measures between nodes. There are 2 types of plots, shown in the image below. Both types of plots can be generated as an animation to view changes over time or as standalone plots.
Lines drawn on a 2D representation of a skull |
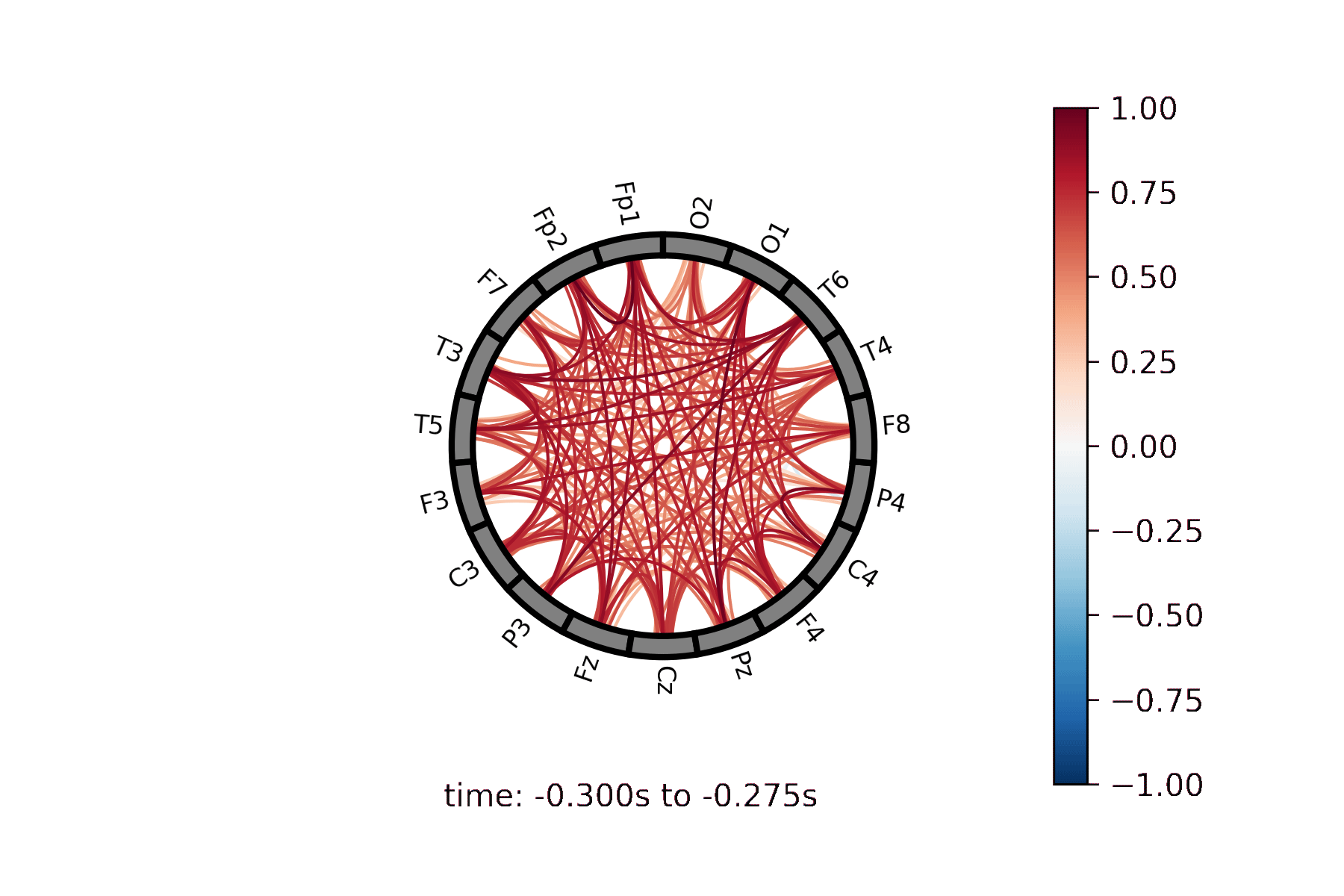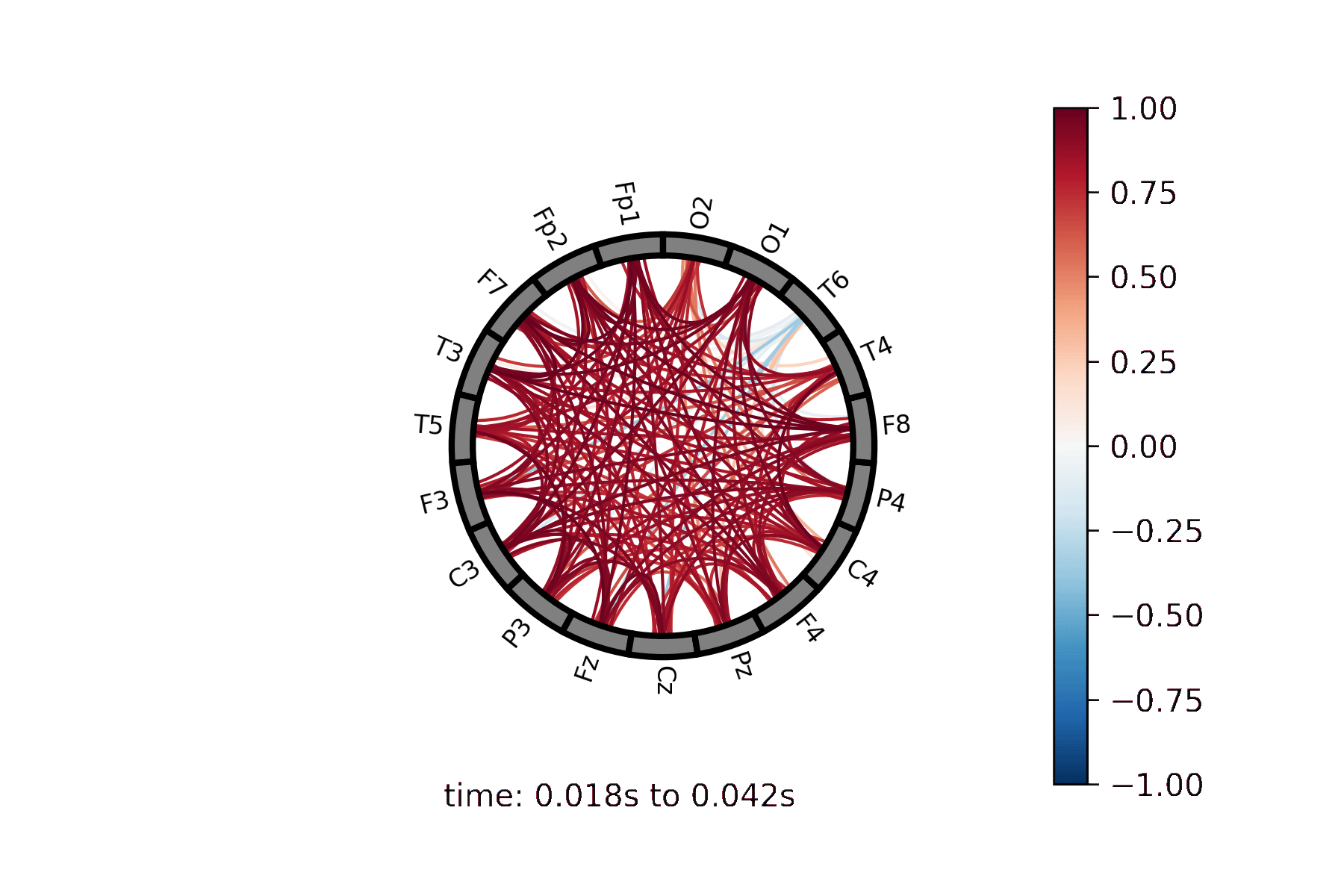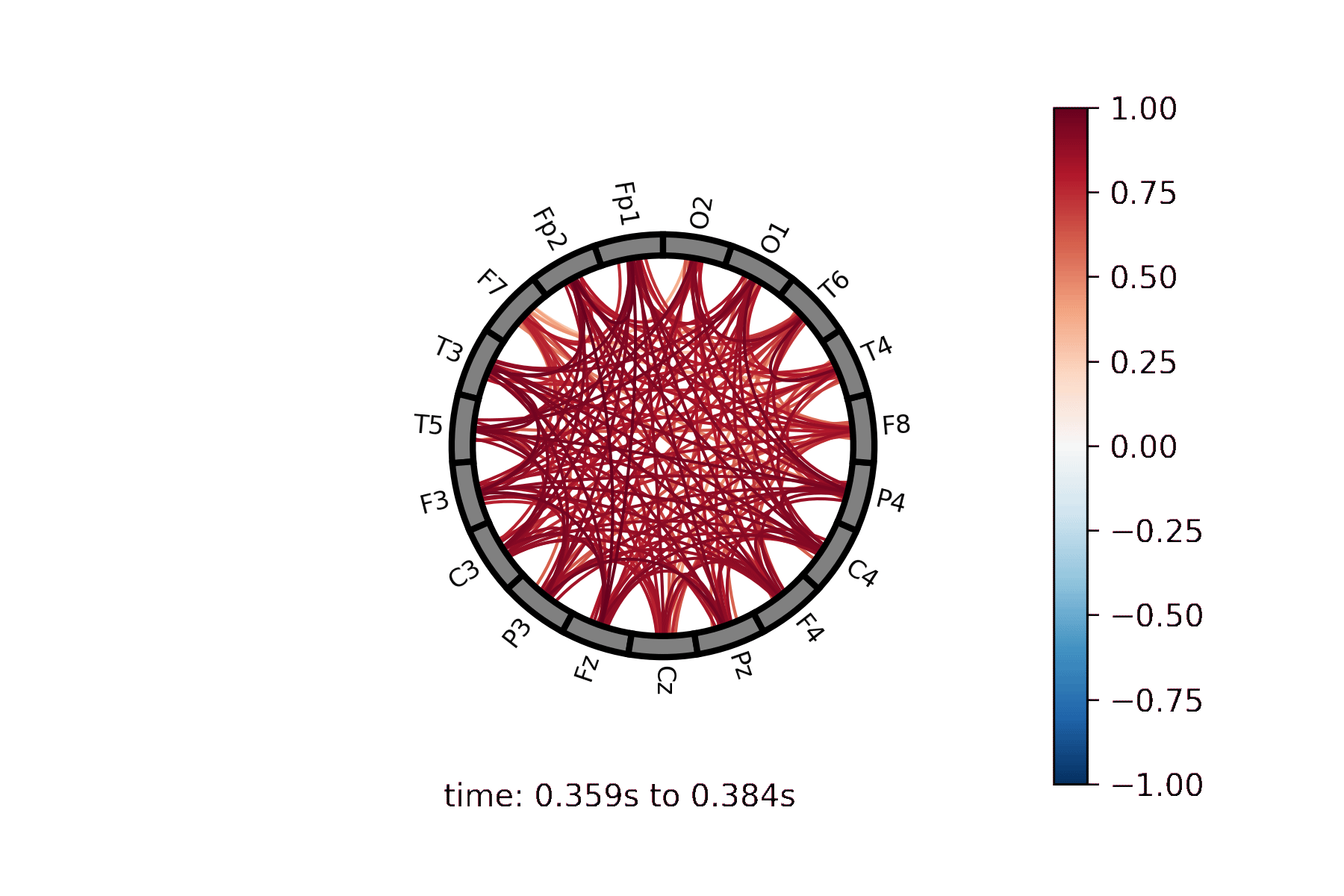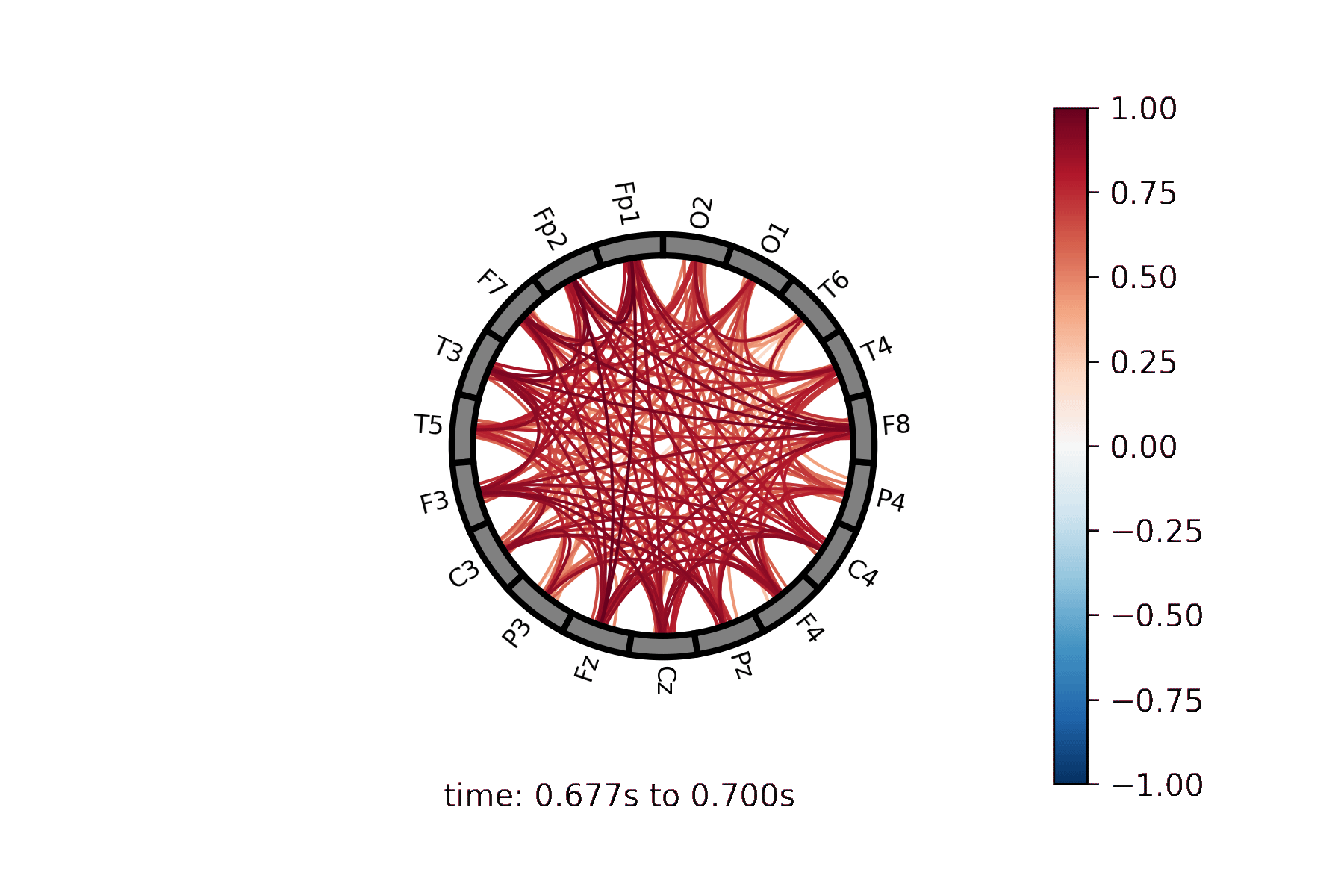
Lines drawn between nodes represented in a circle |
General Setup¶
Import required modules¶
from simpl_eeg import connectivity, eeg_objects
import warnings
warnings.filterwarnings('ignore')
Create epoched data¶
For additional options see Creating EEG Objects section.
experiment_folder = "../../data/927"
nth_epoch = 0
epochs = eeg_objects.Epochs(experiment_folder)
epoch = epochs.get_epoch(nth_epoch)
Reading /Users/mpin/Documents/MDS/capstone/simpl_eeg_capstone/data/927/fixica.fdt
loaded raw from ../../data/927/fixica.set
Not setting metadata
Not setting metadata
33 matching events found
Setting baseline interval to [-0.2998046875, 0.0] sec
Applying baseline correction (mode: mean)
0 projection items activated
Loading data for 33 events and 2049 original time points ...
0 bad epochs dropped
Create a Connectivity Plot animation¶
Define parameters¶
A detailed description of all parameters can be found in the connectivity.animate_connectivity docstring:
help(connectivity.animate_connectivity)
Help on function animate_connectivity in module simpl_eeg.connectivity:
animate_connectivity(epoch, calc_type='correlation', steps=20, pair_list=[], threshold=0, show_sphere=True, colormap='RdBu_r', vmin=None, vmax=None, line_width=None, title=None, colorbar=True, timestamp=True, frame_rate=12.0, **kwargs)
Animate 2d EEG nodes on scalp with lines representing connectivity
Args:
epochs: mne.epochs.Epochs
Epoch to visualize
calc_type: str (optional)
Connectivity calculation type. Defaults to "correlation".
steps: int (optional)
Number of frames to use in correlation caluclation. Defaults to 20.
pair_list: [str] (optional)
List of node pairs. Defaults to [], which indicates all pairs.
threshold: int (optional)
Unsigned connectivity threshold to display connection.
Defaults to 0.
show_sphere: bool (optional)
Whether to show the cartoon head or not. Defaults to True.
colormap: str (optional)
Colour scheme to use. Defaults to "RdBu_r"
vmin: int (optional)
The minimum for the scale. Defaults to None.
vmin: int (optional)
The maximum for the scale. Defaults to None.
line_width: int (optional)
The line width for the connections.
Defaults to None for non-static width.
title: str (optional)
The title to display on the plot. Defaults to None for no title.
colorbar: bool (optional)
Whether to show the colorbar. Defaults to True.
timestamp: bool (optional)
Whether to show the timestamp caption. Defaults to True.
frame_rate: int or float (optional)
The frame rate to genearte the final animation with. Defaults to 12.0.
**kwargs: dict (optional)
Optional arguments to pass to mne.viz.plot_sensors()
Full list of options available at
https://mne.tools/stable/generated/mne.viz.plot_sensors.html
Returns:
matplotlib.animation.Animation:
Animation of connectivity plot
vmin = -1
vmax = 1
colormap = "RdBu_r"
calc_type = "correlation"
line_width = None
steps = 50
threshold = 0
show_sphere = True
PAIR_OPTIONS = {
"all_pairs": [],
"local_anterior": "Fp1-F7, Fp2-F8, F7-C3, F4-C4, C4-F8, F3-C3",
"local_posterior": "T5-C3, T5-O1, C3-P3, C4-P4, C4-T6, T6-O2",
"far_coherence": "Fp1-T5, Fp2-T6, F7-T5, F7-P3, F7-O1, T5-F3, F3-P3, F4-P4, P4-F8, F8-T6, F8-O2, F4-T6",
"prefrontal_to_frontal_and_central": "Fp1-F3, Fp1-C3, Fp2-F4, Fp2-C4",
"occipital_to_parietal_and_central": "C3-O1, P3-O1, C4-O2, P4-O4",
"prefrontal_to_parietal": "Fp1-P3, Fp2-P4",
"frontal_to_occipital": "F3-O1, P4-O2",
"prefrontal_to_occipital": "Fp1-O1, Fp2-O2"
}
# select from the PAIR_OPTIONS options above or use a custom pair.
pair_list = [] # leave as an empty list if you want all pairs
# example of referencing a pair from the list
pair_list = PAIR_OPTIONS["far_coherence"]
Generating the animation¶
# for displaying animation in jupyter notebook
%matplotlib inline
%%capture
anim = connectivity.animate_connectivity(
epoch,
calc_type=calc_type,
steps=steps,
pair_list=pair_list,
threshold=threshold,
show_sphere=show_sphere,
colormap=colormap,
vmin=vmin,
vmax=vmax,
line_width=line_width,
)
from IPython.display import HTML
html_plot = anim.to_jshtml()
video = HTML(html_plot)
video
Saving the animation¶
Save as html¶
html_file_path = "examples/connectivity.html"
html_file = open(html_file_path, "w")
html_file.write(html_plot)
html_file.close()
Save as gif¶
gif_file_path = "examples/connectivity.gif"
# set frames per second (fps) and resolution (dpi)
anim.save(gif_file_path, fps=3, dpi=300)
Save as mp4¶
mp4_file_path = "examples/connectivity.mp4"
anim.save(mp4_file_path, fps=3, dpi=300)
Note
If FFMpegWriter does not work on your computer you can save the file as a gif first and then convert it into mp4 file by running the code below.
import moviepy.editor as mp
clip = mp.VideoFileClip(gif_file_path)
clip.write_videofile(mp4_file_path)
Create a Connectivity Circle animation¶
Define parameters¶
A detailed description of all parameters can be found in the connectivity.animate_connectivity_circle docstring:
help(connectivity.animate_connectivity_circle)
Help on function animate_connectivity_circle in module simpl_eeg.connectivity:
animate_connectivity_circle(epoch, calc_type='correlation', max_connections=50, steps=20, colormap='RdBu_r', vmin=None, vmax=None, line_width=1.5, title=None, colorbar=True, timestamp=True, frame_rate=12.0, **kwargs)
Animate connectivity circle
Args:
epoch: mne.epochs.Epochs
Epoch to visualize
calc_type: str (optional)
Connectivity calculation type. Defaults to "correlation".
max_connections: int (optional)
Number of connections to display. Defaults to 50.
steps: int (optional)
Number of frames to use in correlation caluclation. Defaults to 20.
colormap: str (optional)
Colour scheme to use. Defaults to "RdBu_r".
vmin: int (optional)
The minimum for the scale. Defaults to None.
vmin: int (optional)
The maximum for the scale. Defaults to None.
line_width: int (optional)
The line width for the connections. Defaults to 1.5.
title: str (optional)
The title to display on the plot. Defaults to None for no title.
colorbar: bool (optional)
Whether to display the colorbar or not. Defaults to True.
timestamp: bool (optional)
Whether to display the timestamp caption. Defaults to True.
frame_rate: int or float (optional)
The frame rate to genearte the final animation with. Defaults to 12.0.
**kwargs: dict (optional)
Optional arguments to pass to mne.viz.plot_connectivity_circle()
Full list of options available at
https://mne.tools/stable/generated/mne.viz.plot_connectivity_circle.html
Returns:
matplotlib.animation.Animation:
Animation of connectivity plot
vmin = -1
vmax = 1
colormap = "RdBu_r"
calc_type = "correlation"
line_width = 1
steps = 50
max_connections = 50
Generating the animation¶
# for displaying animation in jupyter notebook
%matplotlib notebook
%%capture
anim = connectivity.animate_connectivity_circle(
epoch,
calc_type=calc_type,
max_connections=max_connections,
steps=steps,
colormap=colormap,
vmin=vmin,
vmax=vmax,
line_width=line_width,
)
from IPython.display import HTML
html_plot = anim.to_jshtml()
video = HTML(html_plot)
video
Saving the animation¶
Save as html¶
html_file_path = "examples/connectivity_circle.html"
html_file = open(html_file_path, "w")
html_file.write(html_plot)
html_file.close()
Save as gif¶
gif_file_path = "examples/connectivity_circle.gif"
# set frames per second (fps) and resolution (dpi)
anim.save(gif_file_path, fps=3, dpi=300)
Save as mp4¶
mp4_file_path = "examples/connectivity_cicle.mp4"
anim.save(mp4_file_path, fps=3, dpi=300)
Note
If FFMpegWriter does not work on your computer you can save the file as a gif first and then convert it into mp4 file by running the code below.
import moviepy.editor as mp
clip = mp.VideoFileClip(gif_file_path)
clip.write_videofile(mp4_file_path)
Create a Connectivity Plot¶
Define parameters¶
A detailed description of all animation parameters can be found in the connectivity.plot_connectivity docstring:
help(connectivity.plot_connectivity)
Help on function plot_connectivity in module simpl_eeg.connectivity:
plot_connectivity(epoch, fig=None, locations=None, calc_type='correlation', pair_list=[], threshold=0, show_sphere=True, readjust_sphere='auto', colormap='RdBu_r', vmin=None, vmax=None, line_width=None, title=None, colorbar=True, caption=None, **kwargs)
Plot 2d EEG nodes on scalp with lines representing connectivity
Args:
epoch: mne.epochs.Epochs
Epoch to visualize
fig: matplotlib.pyplot.figure (optional)
Figure to plot on. Defaults to None.
locations: [matplotlib.text.Text] (optional)
List of node locations. Defaults to None.
calc_type: str
Connectivity calculation type
pair_list: [str] (optional)
List of node pairs. Defaults to [], which indicates all pairs.
threshold: int (optional)
Unsigned connectivity threshold to display connection.
Defaults to 0.
show_sphere: bool (optional)
Whether to show the cartoon head or not. Defaults to True.
readjust_sphere: bool or "auto" (optional)
Tries to re-align cartoon head but may overcorrect. Defaults to "auto".
colormap: str (optional)
Colour scheme to use. Defaults to "RdBlu_r".
vmin: int (optional)
The minimum for the scale. Defaults to None.
vmin: int (optional)
The maximum for the scale. Defaults to None.
line_width: int (optional)
The line width for the connections.
Defaults to None for non-static width.
title: str (optional)
The title to display on the plot. Defaults to None for no title.
colorbar: bool (optional)
Whether to display the colorbar. Defaults to True.
caption: str (optional)
The caption to display at the bottom of the plot. Defaults to None.
**kwargs: dict (optional)
Optional arguments to pass to mne.viz.plot_sensors()
Full list of options available at
https://mne.tools/stable/generated/mne.viz.plot_sensors.html
Returns:
matplotlib.pyplot.figure:
The generated connectivity figure
vmin = -1
vmax = 1
colormap = "RdBu_r"
calc_type = "correlation"
line_width = None
threshold = 0
show_sphere = True
readjust_sphere = 'auto'
PAIR_OPTIONS = {
"all_pairs": [],
"local_anterior": "Fp1-F7, Fp2-F8, F7-C3, F4-C4, C4-F8, F3-C3",
"local_posterior": "T5-C3, T5-O1, C3-P3, C4-P4, C4-T6, T6-O2",
"far_coherence": "Fp1-T5, Fp2-T6, F7-T5, F7-P3, F7-O1, T5-F3, F3-P3, F4-P4, P4-F8, F8-T6, F8-O2, F4-T6",
"prefrontal_to_frontal_and_central": "Fp1-F3, Fp1-C3, Fp2-F4, Fp2-C4",
"occipital_to_parietal_and_central": "C3-O1, P3-O1, C4-O2, P4-O4",
"prefrontal_to_parietal": "Fp1-P3, Fp2-P4",
"frontal_to_occipital": "F3-O1, P4-O2",
"prefrontal_to_occipital": "Fp1-O1, Fp2-O2"
}
# select from the PAIR_OPTIONS options above or use a custom pair.
pair_list = [] # leave as an empty list if you want all pairs
# example of referencing a pair from the list
pair_list = PAIR_OPTIONS["far_coherence"]
Generating a standalone plot¶
Note
Generating a plot will use the first frame in the epoch, so make sure to update your epoch object to contain your frame of interest.
# for displaying plot in jupyter notebook
%matplotlib inline
%%capture
plot = connectivity.plot_connectivity(
epoch,
)
plot
Saving the plot¶
You can change the file to different formats by changing the format argument in the function. It supports png, pdf, svg.
file_path = "examples/connectivity.svg"
plot.figure.savefig(file_path)
Create a Connectivity Circle Plot¶
Define parameters¶
A detailed description of all animation parameters can be found in the connectivity.plot_conn_circle docstring:
help(connectivity.plot_conn_circle)
Help on function plot_conn_circle in module simpl_eeg.connectivity:
plot_conn_circle(epoch, fig=None, calc_type='correlation', max_connections=50, ch_names=[], colormap='RdBu_r', vmin=None, vmax=None, line_width=1.5, title=None, colorbar=True, caption=None, **kwargs)
Plot connectivity circle
Args:
epoch: mne.epochs.Epochs
Epoch to visualize
fig: matplotlib.pyplot.figure (optional)
Figure to plot on. Defaults to None.
calc_type: str (optional)
Connectivity calculation type. Defaults to "correlation"
max_connections: int (optional)
Maximum connections to plot. Defaults to 50.
ch_names: [str] (optional)
List of channel names to display.
Defaults to [], which indicates all channels.
vmin: int (optional)
The minimum for the scale. Defaults to None.
vmin: int (optional)
The maximum for the scale. Defaults to None.
colormap: str (optional)
Colour scheme to use. Defaults to "RdBu_r".
colormap: bool (optional)
Whether to plot the colorbar. Defaults to True.
line_width: int (optional)
The line width for the connections. Defaults to 1.5.
title: str (optional)
The title to display on the plot. Defaults to None for no title.
colorbar: bool (optional)
Whether to display the colorbar or not. Defaults to True.
caption: str (optional)
The caption to display at the bottom of the plot. Defaults to None.
**kwargs: dict (optional)
Optional arguments to pass to mne.viz.plot_connectivity_circle()
Full list of options available at
https://mne.tools/stable/generated/mne.viz.plot_connectivity_circle.html
Returns:
matplotlib.pyplot.figure:
Connectivity circle figure
vmin = -1
vmax = 1
colormap = "RdBu_r"
calc_type = "correlation"
line_width = 1
max_connections = 50
Generating a standalone plot¶
Note
Generating a plot will use the first frame in the epoch, so make sure to update your epoch object to contain your frame of interest.
# for displaying plot in jupyter notebook
%matplotlib inline
%%capture
plot = connectivity.plot_conn_circle(
epoch,
)
plot
Saving the plot¶
You can change the file to different formats by changing the format argument in the function. It supports png, pdf, svg.
file_path = "examples/connectivity_circle.svg"
plot.figure.savefig(file_path)