3D Brain Visualizations¶
3D topographic brain map¶
The 3D topographic brain map provides a view of voltage measurements as a heatmap converted to estimated position on the brain. There are 2 plot options with different backends:
matplotlib (the version found on the UI)
pyVista (a better rendering but incompatible with the UI)
Both plots can be generated as an animation to view changes over time or as a standalone plot.
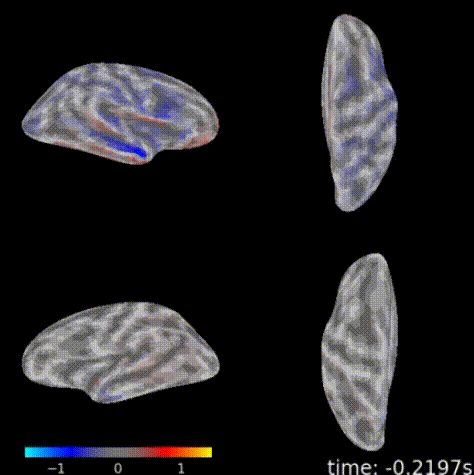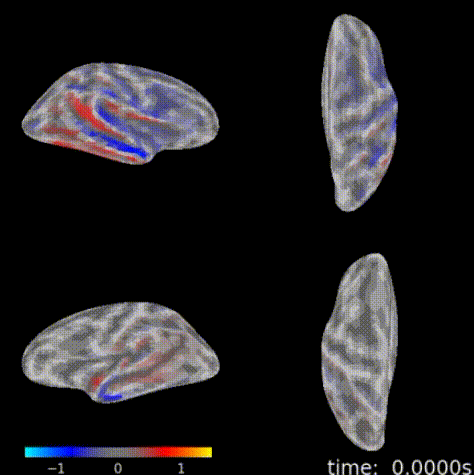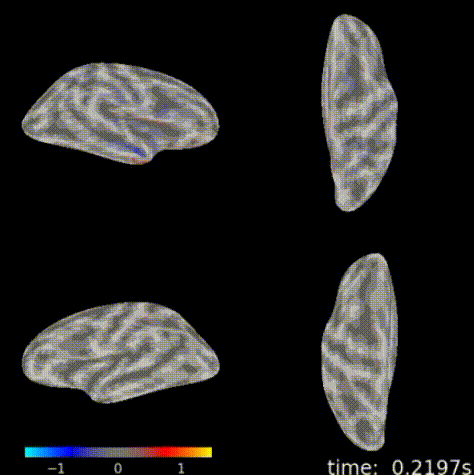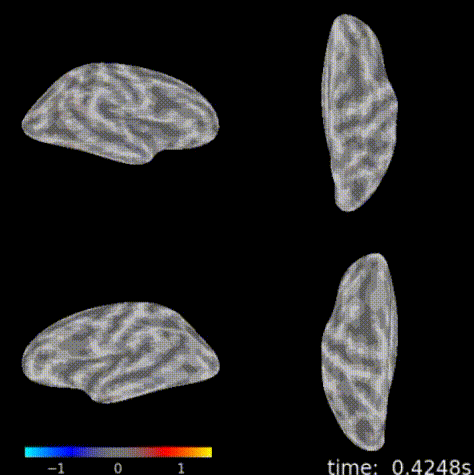
General Setup¶
Import required modules¶
from simpl_eeg import topomap_3d_brain, eeg_objects
Create epoched data¶
For additional options see Creating EEG Objects section.
experiment_folder = "../../data/109"
epochs = eeg_objects.Epochs(experiment_folder)
frame_steps = 100
epoch = epochs.skip_n_steps(frame_steps)
Reading /Users/mpin/Documents/MDS/capstone/simpl_eeg_capstone/data/109/fixica.fdt
/Users/mpin/Documents/MDS/capstone/simpl_eeg_capstone/simpl_eeg/eeg_objects.py:199: RuntimeWarning: Data file name in EEG.data (109 whole fixed ica.fdt) is incorrect, the file name must have changed on disk, using the correct file name (fixica.fdt).
raw = mne.io.read_raw_eeglab(data_path)
loaded raw from ../../data/109/fixica.set
Not setting metadata
Not setting metadata
33 matching events found
Setting baseline interval to [-0.2998046875, 0.0] sec
Applying baseline correction (mode: mean)
0 projection items activated
Loading data for 33 events and 2049 original time points ...
0 bad epochs dropped
/Users/mpin/Documents/MDS/capstone/simpl_eeg_capstone/simpl_eeg/eeg_objects.py:823: RuntimeWarning: The measurement information indicates a low-pass frequency of 1024 Hz. The decim=100 parameter will result in a sampling frequency of 20.48 Hz, which can cause aliasing artifacts.
return epochs.copy().decimate(num_steps)
Generate forward and inverse (optional)¶
Note
Before an animation or plot can be generated, an “inverse” (abbreviated as “stc”) must first be generated. An inverse can be generated from an epoch and a “forward” (abbreviated as “fwd”), which itself can be generated from an epoch alone. If only an epoch is provided to either of the plotting functions the stc and fwd will be automatically generated, HOWEVER this will increase the time it takes to generate the figure.
The forward/inverse are used to retrieve a brain model to attach the EEG data and to do some of the mapping calculations. The forward downloads ‘fsaverage’ MRI data which represents a brain averaged out from dozens of different patients.
You may pass one of three combinations to generate a figure in either of the plotting functions.
An ‘stc’ (fastest)
An ‘epoch’ AND a ‘fwd’ (fast)
An ‘epoch’ (slow)
Generate Forward¶
fwd = topomap_3d_brain.create_fsaverage_forward(epoch)
0 files missing from root.txt in /Users/mpin/mne_data/MNE-fsaverage-data
0 files missing from bem.txt in /Users/mpin/mne_data/MNE-fsaverage-data/fsaverage
Source space : /Users/mpin/mne_data/MNE-fsaverage-data/fsaverage/bem/fsaverage-ico-5-src.fif
MRI -> head transform : /Users/mpin/opt/miniconda3/lib/python3.8/site-packages/mne/data/fsaverage/fsaverage-trans.fif
Measurement data : instance of Info
Conductor model : /Users/mpin/mne_data/MNE-fsaverage-data/fsaverage/bem/fsaverage-5120-5120-5120-bem-sol.fif
Accurate field computations
Do computations in head coordinates
Free source orientations
Reading /Users/mpin/mne_data/MNE-fsaverage-data/fsaverage/bem/fsaverage-ico-5-src.fif...
Read 2 source spaces a total of 20484 active source locations
Coordinate transformation: MRI (surface RAS) -> head
0.999994 0.003552 0.000202 -1.76 mm
-0.003558 0.998389 0.056626 31.09 mm
-0.000001 -0.056626 0.998395 39.60 mm
0.000000 0.000000 0.000000 1.00
Read 19 EEG channels from info
Head coordinate coil definitions created.
Source spaces are now in head coordinates.
Setting up the BEM model using /Users/mpin/mne_data/MNE-fsaverage-data/fsaverage/bem/fsaverage-5120-5120-5120-bem-sol.fif...
Loading surfaces...
Loading the solution matrix...
Three-layer model surfaces loaded.
Loaded linear_collocation BEM solution from /Users/mpin/mne_data/MNE-fsaverage-data/fsaverage/bem/fsaverage-5120-5120-5120-bem-sol.fif
Employing the head->MRI coordinate transform with the BEM model.
BEM model fsaverage-5120-5120-5120-bem-sol.fif is now set up
Source spaces are in head coordinates.
Checking that the sources are inside the surface and at least 5.0 mm away (will take a few...)
Skipping interior check for 2433 sources that fit inside a sphere of radius 47.7 mm
Skipping solid angle check for 0 points using Qhull
Skipping interior check for 2241 sources that fit inside a sphere of radius 47.7 mm
Skipping solid angle check for 0 points using Qhull
Setting up for EEG...
Computing EEG at 20484 source locations (free orientations)...
Finished.
Generate Inverse¶
stc = topomap_3d_brain.create_inverse_solution(epoch, fwd)
Computing rank from data with rank=None
Using tolerance 1.4e-12 (2.2e-16 eps * 19 dim * 3.4e+02 max singular value)
Estimated rank (eeg): 19
EEG: rank 19 computed from 19 data channels with 0 projectors
Reducing data rank from 19 -> 19
/Users/mpin/Documents/MDS/capstone/simpl_eeg_capstone/simpl_eeg/topomap_3d_brain.py:282: RuntimeWarning: Too few samples (required : 100 got : 21), covariance estimate may be unreliable
noise_cov = mne.compute_covariance(epoch, method=covariance_method)
Estimating covariance using EMPIRICAL
Done.
Estimating covariance using SHRUNK
Done.
Using cross-validation to select the best estimator.
Number of samples used : 21
log-likelihood on unseen data (descending order):
shrunk: -65.605
empirical: -257.367
selecting best estimator: shrunk
[done]
Converting forward solution to surface orientation
No patch info available. The standard source space normals will be employed in the rotation to the local surface coordinates....
Converting to surface-based source orientations...
[done]
Computing inverse operator with 19 channels.
19 out of 19 channels remain after picking
Selected 19 channels
Creating the depth weighting matrix...
19 EEG channels
limit = 20485/20484 = 9.583809
scale = 9.50316e+09 exp = 0.8
Applying loose dipole orientations to surface source spaces: 0.2
Whitening the forward solution.
Computing rank from covariance with rank=None
Using tolerance 2.4e-13 (2.2e-16 eps * 19 dim * 56 max singular value)
Estimated rank (eeg): 19
EEG: rank 19 computed from 19 data channels with 0 projectors
Setting small EEG eigenvalues to zero (without PCA)
Creating the source covariance matrix
Adjusting source covariance matrix.
Computing SVD of whitened and weighted lead field matrix.
largest singular value = 2.54939
scaling factor to adjust the trace = 6.64201e+24 (nchan = 19 nzero = 0)
Preparing the inverse operator for use...
Scaled noise and source covariance from nave = 1 to nave = 1
Created the regularized inverter
The projection vectors do not apply to these channels.
Created the whitener using a noise covariance matrix with rank 19 (0 small eigenvalues omitted)
Computing noise-normalization factors (dSPM)...
/Users/mpin/Documents/MDS/capstone/simpl_eeg_capstone/simpl_eeg/topomap_3d_brain.py:284: RuntimeWarning: No average EEG reference present in info["projs"], covariance may be adversely affected. Consider recomputing covariance using with an average eeg reference projector added.
inverse_operator = make_inverse_operator(epoch.info, fwd, noise_cov,
/Users/mpin/Documents/MDS/capstone/simpl_eeg_capstone/simpl_eeg/topomap_3d_brain.py:284: RuntimeWarning: No average EEG reference present in info["projs"], covariance may be adversely affected. Consider recomputing covariance using with an average eeg reference projector added.
inverse_operator = make_inverse_operator(epoch.info, fwd, noise_cov,
[done]
Picked 19 channels from the data
Computing inverse...
Eigenleads need to be weighted ...
Processing epoch : 1 / 1
[done]
Create a matplotlib 3D brain animation¶
Simple Plot (+ auto-generating forward and inverse)¶
Define parameters (simple)¶
A detailed description of all parameters can be found in the topomap_3d_brain.animate_matplot_brain docstring:
help(topomap_3d_brain.animate_matplot_brain)
Help on function animate_matplot_brain in module simpl_eeg.topomap_3d_brain:
animate_matplot_brain(epoch=None, fwd='auto', stc='auto', views=['lat', 'dor', 'fro'], size=200, hemi='both', colormap='mne', colorbar=True, colormap_limit_type='lims', vmin=None, vmid=None, vmax=None, spacing='oct5', smoothing_steps=3, timestamp=True, frame_rate=12, **kwargs)
Creates an animated view of all timestamp observations an mne.epochs.Epochs data using a matplotlib backend.
If multiple views are used then speed becomes significantly slower. Colorbar placement may be inconsistent.
Parameters:
epoch: mne.epochs.Epoch or None
MNE epochs object containing portions of raw EEG data built around specified
timestamp(s). If no fwd and stc are provided a fwd will be generated from the
epoch using create_fsaverage_forward(). The epoch and fwd (either provided or
generated) will then be used in create_inverse_solution() to generate an stc that
the brain figure will be generated from.
fwd: mne.forward.forward.Forward or 'auto'
MNE forward object. If provided alongside an epoch they will both be used in
create_fsaverage_forward() to create an stc which the brain figure will then be
generated from. Defaults to 'auto'.
stc: mne.source_estimate.SourceEstimate or 'auto'
'inverse_solution' to generate the plot from. If set to "auto" then an stc will be
automatically generated from either an epoch or an epoch and a fwd, however, this
will significantly slow down rendering time. Defaults to 'auto'.
views: str or list
Specifies the 'view' parameter in the mne.SourceEstimate.plot() function. For any backend
can be any combination of 'lat' (lateral), 'med' (medial), 'ros' (rostral), 'cau' (caudal),
'dor' (dorsal), 'ven'(ventral), 'fro'(frontal), 'par' (parietal). The following arguments
are also accepted but are NOT compatible with the matplotlib backend 'axi' (axial), 'sag'
(sagittal), and 'cor'(coronal). Defaults to ['lat', 'fro', 'dor'].
size: int
Size will be divided by 100 and rounded the closest inch which will then be used as the
height per view. For example, entering 100 will result in 1 inch per view. If plotting
multiple views overall size of the multiplot is automatically calculated to fit all
views. Defaults to 200.
hemi: 'lh’ or ‘rh’ or ‘both’ or ‘split’
Specifies the 'initial_time' parameter in the mne.SourceEstimate.plot() function. Can be
one of ‘lh’, ‘rh’, ‘both’, or ‘split’. Defaults to 'both'. Note that 'split' and 'both' will
return a 'split' view since both is not avalible with a matplotlib backend. Defaults to 'both'.
colormap: str or np.ndarray of float, shape(n_colors, 3 | 4)
Specifies the 'colormap' parameter in the mne.SourceEstimate.plot() function. Can use a
matplotlib colormap by name or take a custom look up table as input. Defaults to "mne".
colorbar: bool
Determines whether to include a colorbar on the plot not. Defaults to True.
colormap_limit_type: str
Can be either "lims" or "pos_lims". "lims" means that your vmin, vmid, and vmax values will specify the
"Lower, middle, and upper bounds for colormap". Using "pos_lims" will lead to vmin, vmid, and vmax representing
the "Lower, middle, and upper bound for colormap. Positive values will be mirrored directly across
zero during colormap construction to obtain negative control points." Defaults to "lims".
vmin: float
Specifies the lower value of the colormap limit. If no value is specified then
limits will be automatically calculated based on the mne.SourceEstimate.plot() function defaults OR
will be the negative value of vmax if only that is provided.
vmid: float
Specifies the middle value of the colormap limit. If no value is specified then
limits will be automatically calculated based on the mne.SourceEstimate.plot() function defaults OR
will be the value between vmin and vmax if one/both of them is provided.
vmax: float
Specifies the middle value of the colormap limit. If no value is specified then
limits will be automatically calculated based on the mne.SourceEstimate.plot() function defaults OR
will be the negative value of vmin if only that is provided.
spacing: str
Specifies the 'spacing' parameter in the mne.SourceEstimate.plot() function. "The spacing to use for the
source space. Can be 'ico#' for a recursively subdivided icosahedron, 'oct#' for a recursively subdivided
octahedron. In general, you can speed up the plotting by selecting a sparser source
space. Has no effect with mayavi backend. Defaults to ‘oct6’".
smoothing_steps: int
Specifies the 'smoothing_steps' parameter in the mne.SourceEstimate.plot() function. "The amount of smoothing".
Defaults to 3.
timestamp: bool
Specifies whether or not to show the timestamp on the plot relative to the time in the epoch that
is being shown. Defaults to True.
frame_rate: int or float
The frame rate to render the animation at. Defautls to 12.
Returns:
matplotlib.animation.FuncAnimation:
Animation containing frames from all of the avalible times in the passed in epoch.
Define parameters¶
# change values below to values of interest
# arguments built into the package
stc = stc
views=['lat', 'dor']
size=200
hemi='both'
colormap='mne'
colorbar=True
colormap_limit_type='lims'
vmin=-2
vmax=2
spacing='oct5'
smoothing_steps=5
timestamp=True
frame_rate=12
# some useful (and tested) arguments from the
# MNE.viz.plot_topomap function (see **kwargs) include...
surface='inflated'
cortex='classic'
Note
Remember there are three options for passing a combination of epoch, fwd, and stc to generate a plot. These are… 1. stc=stc
epoch=epoch, fwd=fwd
epoch=epoch
Generate animation with matplotlib backend¶
%%capture
matplotlib_animation = topomap_3d_brain.animate_matplot_brain(
stc=stc,
views=views,
hemi=hemi,
colormap=colormap,
colorbar=colorbar,
colormap_limit_type=colormap_limit_type,
vmin=vmin,
vmax=vmax,
spacing=spacing,
smoothing_steps=smoothing_steps,
timestamp=timestamp,
frame_rate=frame_rate,
surface=surface,
cortex=cortex
)
from IPython.display import HTML
video = HTML(matplotlib_animation.to_jshtml())
video
Saving the animation¶
Save as gif¶
anim_brain = topomap_3d_brain.animate_matplot_brain(stc = stc, views = 'lat', hemi = 'lh')
gif_file_path = "examples/topomap_3d_brain.gif"
anim_brain.save(gif_file_path, fps=5, dpi=300)
Save as mp4¶
mp4_file_path = "examples/topo_2d.mp4"
anim_brain.save(mp4_file_path, fps=5, dpi=300)
Note
If FFMpegWriter does not work on your computer you can save the file as a gif first and then convert it into mp4 file by running the code below.
import moviepy.editor as mp
clip = mp.VideoFileClip(gif_file_path)
clip.write_videofile(mp4_file_path)
Create a matplotlib 3D brain figure¶
Generating a matplotlib plot¶
Define parameters¶
A detailed description of all animation parameters can be found in the topomap_3d_brain.plot_topomap_3d_brain docstring:
help(topomap_3d_brain.plot_topomap_3d_brain)
%%capture
matplot_brain_fig = topomap_3d_brain.plot_topomap_3d_brain(stc=stc,
recording_number=2,
hemi='lh',
views=['lat'],
vmin=-2,
vmax=2)
matplot_brain_fig
Save the plot¶
You can change the file to different formats by changing the format argument in the function. It supports png, pdf, svg.
file_path = "examples/topomap_3d_brain.svg"
matplot_brain_fig.savefig(file_path, format='svg')
Create a pyVista 3D brain animation¶
Note
The dependencies for the following functions are not included with the base instalation of the simpl_eeg package due to compatability issues with certain operating systems. If you wish to use them please install ‘PyVista’, ‘PyQt5’, ‘pyvistaqt’, and ‘pyqt5-qt5’ through your preferred package installation method https://mne.tools/stable/install/mne_python.html.
Generate figure with pyvista backend¶
pyvista_brain_fig = topomap_3d_brain.plot_topomap_3d_brain(stc = stc, backend = 'pyvista')
Save animation with pyvista backend¶
topomap_3d_brain.save_animated_topomap_3d_brain(pyvista_brain_fig, filename = "brain_animation.gif")
