2D Head Visualizations¶
2D topographic head map¶
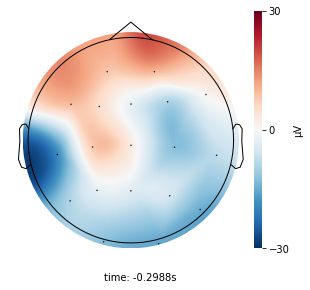
The 2D topographic head map provides a view of voltage measurements as a heatmap imposed on an image of a 2D skull. It can be generated as an animation to view changes over time or as a standalone plot.
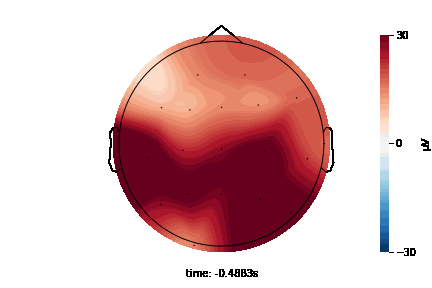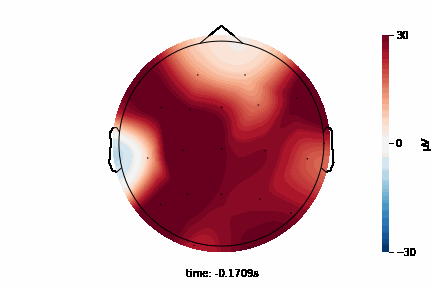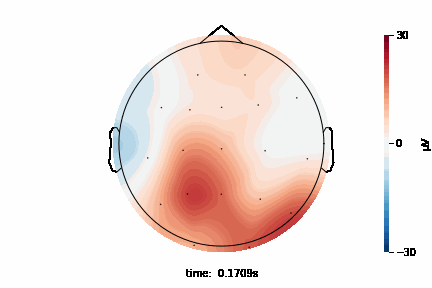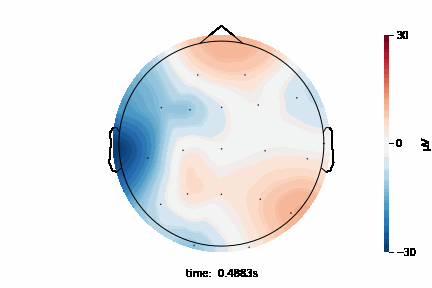
General Setup¶
Import required modules¶
from simpl_eeg import topomap_2d, eeg_objects
Create epoched data¶
For additional options see Creating EEG Objects section.
experiment_folder = "../../data/109"
nth_epoch = 0
num_steps = 50 # number of steps to skip to shorten epoch
epochs = eeg_objects.Epochs(experiment_folder)
epoch = epochs.get_epoch(nth_epoch)
shortened_epoch = epochs.skip_n_steps(num_steps)
Reading /Users/mpin/Documents/MDS/capstone/simpl_eeg_capstone/data/109/fixica.fdt
/Users/mpin/Documents/MDS/capstone/simpl_eeg_capstone/simpl_eeg/eeg_objects.py:199: RuntimeWarning: Data file name in EEG.data (109 whole fixed ica.fdt) is incorrect, the file name must have changed on disk, using the correct file name (fixica.fdt).
raw = mne.io.read_raw_eeglab(data_path)
loaded raw from ../../data/109/fixica.set
Not setting metadata
Not setting metadata
33 matching events found
Setting baseline interval to [-0.2998046875, 0.0] sec
Applying baseline correction (mode: mean)
0 projection items activated
Loading data for 33 events and 2049 original time points ...
0 bad epochs dropped
/Users/mpin/Documents/MDS/capstone/simpl_eeg_capstone/simpl_eeg/eeg_objects.py:823: RuntimeWarning: The measurement information indicates a low-pass frequency of 1024 Hz. The decim=50 parameter will result in a sampling frequency of 40.96 Hz, which can cause aliasing artifacts.
return epochs.copy().decimate(num_steps)
Create a 2D topographic animation¶
Define parameters¶
A detailed description of all animation parameters can be found in the topomap_2d.animate_topomap_2d docstring:
help(topomap_2d.animate_topomap_2d)
Help on function animate_topomap_2d in module simpl_eeg.topomap_2d:
animate_topomap_2d(epoch, plotting_data=None, colormap='RdBu_r', mark='dot', vmin=-30, vmax=30, sphere='auto', colorbar=True, timestamp=True, frame_rate=12, **kwargs)
Plots a still image mne.epochs.Epochs EEG data as a 2D topomap using the mne.viz.plot_topomap
function.
Parameters:
epoch: mne.epochs.Epochs or mne.evoked.EvokedArray
MNE epochs object (or evoked array object) containing portions of raw EEG data built
around specified timestamp(s).
plotting_data: numpy.ndarray or None
Array of the EEG data from a measurement (not a time interval) to be plotted should be in
the format (num_channels, num_frames) i.e. multiple rows of values for the timestamps you plan
to plot. If plotting_data is provided the heatmap in the figure will be built from it in
conjunction with the 'info' from the provided epoch (containing information like node names
and locations). If no plotting_data is provided the heatmap will be built from the EEG data
in the epoch instead.
colormap: matplotlib colormap or None
Specifies the 'colormap' parameter to be used in the mne.viz.plot_topomap() and
in colorbar generation. Defaults to 'RdBu_r'.
mark: str
Specifies what kind of marker should be shown for each node on the topomap. Can be one of
'dot', 'r+' (for red +'s), 'channel_name', or 'none'. Defaults to 'dot'.
vmin: float
Specifies the 'vmin' parameter in the mne.viz.plot_topomap() function.
Sets the limits for the colors which will be used on the topomap. Value is
in μV. Defaults to -30.
vmax: float
Specifies the 'vmax' parameter in the mne.viz.plot_topomap() function.
Sets the limits for the colors which will be used on the topomap. Value is
in μV. Defaults to +30.
colorbar: bool
Specifies whether or not to include a colorbar in the animation. Removing will lead to
a marginal decrease in rendering times. Defaults to True.
timestamp: bool
Specifies whether or not to show the timestamp on the plot relative to the time in the epoch that
is being shown. Defaults to True.
frame_rate: int or float
The frame rate to genearte the final animation with. Defaults to 12.
**kwargs: various
Additional arguments from the 'mne.viz.plot_topomap' function
(https://mne.tools/stable/generated/mne.viz.plot_topomap.html) may also be passed. Since this function
is used under-the-hood most of these arguments should work exactly as they do in their original
function. The exact list of avalible arguments and their defaults includes: [contours: 0, sphere: 100,
res: 64, extrapolate: 'head', outlines: 'head, mask: None, mask_params: None, image_interp: 'blinear',
show: False, onselect: None, border: 'mean', ch_type: 'eeg']. The 'axes' argument is not avalible
since it does not work with animations. Some arguments may break this function. The arguments that are
tested and known to work include [res, sphere, extrapolate, outlines, contours, outlines, and border].
Returns:
ani: matplotlib.animation.FuncAnimation
matplotlib funcanimation of a 2d topographic map based on the input epoch data.
# change values below to values of interest
# arguments built into the package
colormap = "Spectral"
mark = "dot"
vmin = -40
vmax = 40
colorbar = True
timestamp = True
frame_rate = 12
# some useful (and tested) arguments from the
# MNE.viz.plot_topomap function (see **kwargs) include...
contours = 0
res = 100
sphere = 100
extrapolate = "head"
outlines = "head"
border = "mean"
Generating the animation¶
%%capture
anim = topomap_2d.animate_topomap_2d(
shortened_epoch, # REQUIRED
colormap=colormap,
mark=mark,
vmin=vmin,
vmax=vmax,
colorbar=colorbar,
timestamp=timestamp,
frame_rate=frame_rate,
contours=contours,
res=res,
sphere=sphere,
extrapolate=extrapolate,
outlines=outlines,
border=border
)
from IPython.core.display import HTML
html_plot = anim.to_jshtml()
video = HTML(html_plot)
video
Saving the animation¶
Save as html¶
html_file_path = "examples/topo_2d.html"
html_file = open(html_file_path, "w")
html_file.write(html_plot)
html_file.close()
Save as gif¶
anim = topomap_2d.animate_topomap_2d(shortened_epoch)
gif_file_path = "examples/topo_2d.gif"
anim.save(gif_file_path, fps=5, dpi=300)
Save as mp4¶
anim = topomap_2d.animate_topomap_2d(shortened_epoch)
mp4_file_path = "examples/topo_2d.mp4"
anim.save(mp4_file_path)
Note
If FFMpegWriter does not work on your computer you can save the file as a gif first and then convert it into mp4 file by running the code below.
import moviepy.editor as mp
clip = mp.VideoFileClip(gif_file_path)
clip.write_videofile(mp4_file_path)
Create a 2D topographic plot¶
Define parameters¶
A detailed description of all animation parameters can be found in the topomap_2d.plot_topomap_2d docstring:
help(topomap_2d.plot_topomap_2d)
Help on function plot_topomap_2d in module simpl_eeg.topomap_2d:
plot_topomap_2d(epoch, plotting_data=None, recording_number=0, colormap='RdBu_r', colorbar=True, vmin=-30, vmax=30, mark='dot', sphere='auto', timestamp=True, **kwargs)
Plots a still image mne.epochs.Epochs EEG data as a 2D topomap using the mne.viz.plot_topomap
function.
Parameters:
epoch: mne.epochs.Epochs or mne.evoked.EvokedArray
MNE epochs object (or evoked array object) containing portions of raw EEG data built
around specified timestamp(s).
plotting_data: numpy.ndarray or None
Array of the EEG data from a measurement (not a time interval) to be plotted should be in
the format (num_channels, ) i.e. a single row of values for the only timestamp you plan
to plot. If plotting_data is provided the heatmap in the figure will be built from it in
conjunction with the 'info' from the provided epoch (containing information like node
names and locations). If no plotting_data is provided the heatmap will be built from the
EEG data in the epoch instead. Defaults to None.
recording_number: int
The index of the data recording in the epoch to show in the plot. Defaults to 0.
colormap: matplotlib colormap or None
Specifies the 'colormap' parameter to be used in the mne.viz.plot_topomap() and
in colorbar generation. Defaults to 'RdBu_r'.
colorbar: bool
Specifies whether to include a colorbar or not. Removing it will marginally improve
performance. Defaults to True.
vmin: float
Specifies the 'vmin' parameter in the mne.viz.plot_topomap() function.
Sets the limits for the colors which will be used on the topomap. Value is
in μV. Defaults to -30.
vmax: float
Specifies the 'vmax' parameter in the mne.viz.plot_topomap() function.
Sets the limits for the colors which will be used on the topomap. Value is
in μV. Defaults to +30.
mark: str
Specifies what kind of marker should be shown for each node on the topomap. Can be one of
'dot', 'r+' (for red +'s), 'channel_name', or 'none'. Deaults to 'dot'.
timestamp: bool
Specifies whether or not to show the timestamp on the plot relative to the time in the epoch that
is being shown. Defaults to True.
**kwargs: various
Additional arguments from the 'mne.viz.plot_topomap' function
(https://mne.tools/stable/generated/mne.viz.plot_topomap.html) may also be passed. Since this function
is used under-the-hood most of these arguments should work exactly as they do in their original
function. The exact list of avalible arguments and their defaults includes: [contours: 0, sphere: 100,
res: 64, extrapolate: 'head', outlines: 'head, mask: None, mask_params: None, image_interp: 'blinear',
show: False, onselect: None, border: 'mean', ch_type: 'eeg', axes: None]. Some arguments may break
this function. The arguments that are tested and known to work include [res, sphere, extrapolate,
outlines, contours, outlines, border].
Returns:
topo_2d_fig: matplotlib.image.AxesImage
matplotlib image plot of a 2d topographic map based on the input epoch data.
timestamp=True
recording_number = 2
Generating a standalone plot¶
Note
Generating a plot will use the first data recording in the epoch by default.
plot = topomap_2d.plot_topomap_2d(
epoch,
timestamp=timestamp,
recording_number=recording_number
)
plot;
Saving the plot¶
You can change the file to different formats by changing the format argument in the function. It supports png, pdf, svg.
file_path = "examples/topo_2d.svg"
plot.figure.savefig(file_path)
